Biosynthesis of purine
advertisement

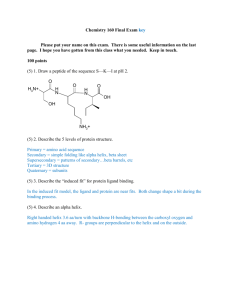
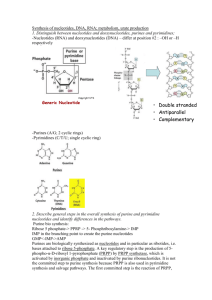
BIOSYNTHESIS OF PURINE 1. INTRODUCTION Nucleotides are essential for many cellular functions, including the storage of genetic information, gene expression, energy metabolism, cell signaling, and biosynthesis. In a cell, nucleotides exist primarily as 5'-triphosphates. ATP is the most prevalent nucleotide, reaching mM concentrations in many cell types, while other nucleotides may be present at much lower concentrations (cAMP). nucleobase Adenine Guanine Thymine Cytosine Uracil Hypoxanthine Xanthine Nucleoside Adenosine Guanosine Thymidine Cytidine Uridine Inosine Xanthosine Nucleotide 5’-monophosphate Adenosine 5’-monophosphate (adenylate, AMP) Guanosine 5’-monophosphate (guanylate, GMP) Thymidine 5’-monophosphate (thymidylate, TMP) Cytidine 5’-monophosphate (cytidylate, CMP) Uridine 5’-monophosphate (uridylate, UMP) Inosine 5’-monophosphate (inosinate, IMP) Xanthosine 5’-monophosphate (xanthylate , XMP) 1 Purine Biosynthesis DE NOVO SYNTHESIS OF PURINES . IMP is synthesized first and serves as a precursor to adenine and guanine nucleotides. Reaction scheme: 2 Step 1: Formation of 5-phosphoribosyl-1-amine from PRPP (committed step) by the action of glutamine phosphoribosyl amidotransferase. PRPP provides the foundation for purine biosynthesis. Glutamine phosphoribosyl amidotransferase has two domains: one hydrolyzes glutamine to ammonia, and the other one is phosphoribosyl transferase domain. Ammonia is channeled to site II without leaving the enzyme. 2- Future N-9 of the purine ring Glu + NH3 Gln + H2O 2- O3PO O3PO CH2 CH2 O -O O -O P O OH NH2 O -configuration of the sugar OP O PPi is replaced by NH2 O inversion of configuration OH PPi OH OH 5-Phosphoribosyl-1-amine 5-phosphoribosyl-1-pyrophosphate (PRPP) This is the committed step. It is feedback inhibited by IMP. GMP, AMP Step 2: Glycine is phosphoribosylamine coupled to the amino group of glycinamide ribonucleotide synthetase ATP + Gly N7 ADP + Pi C4 C5 NH P-ribose P-ribose-NH2 phorphoribosyl amine Gly is coupled to the amino group - O C CH2 C O NH3+ NH3+ CH2 This is the only step in purine biosynthesis that contributes more than one atom to the purine skeleton. glycinamide ribonucleotide O a) The carboxylate group of Gly is activated by phosphorylation to form acylphosphate. O -O P O O- NH3+ C O CH2 3 b) The phosphate is then displaced by the amino group of phophoribosylamine. O O -O P O -O NH3+ C OP-ribose-NH2 P OH O- P-ribose-NH CH2 NH3+ C O CH2 O Step 3: Transfer of a formyl group from N10formyltetrahydrofolate to the amino group of the glycine residue (catalyzed by glycinamide ribonucleotide transferase). N10-formyltetrahydrofolate (THF) serves as a C1 carbon donor. O C8 10 N -formylNH3+ P-ribose NH C THF H NH N7 THF CH2 O C NH P-ribose C4 C H2 N N OH O Formylglycinamide ribonucleotide Glycinamide ribonucleotide CH2 5 N H CH2 C5 N9 -amino terminus is formylated H N N O C H CH2 O N C O- O 10 C N H H2 H2 O CH C C C OH H 10 N -formyl -tetrahydrofolate (THF) Step 4: The inner amide group is converted to an amidine by the replacement with ammonia derived from glutamine: O O H P-ribose NH C ATP H ADP + Pi NH C C C8 NH N7 N9 CH2 P-ribose Glu + NH3 Gln + H2O O Formylglycinamide ribonucleotide NH N3 =NH replaces =O 4 C4 C CH2 C5 N H Formylglycinamidine ribonucleotide Note: Steps 1-4 are catalyzed by a multienzyme complex. Many of the intermediates in purine biosynthesis are unstable in aqueous solution and only exist in solvent protected environment. The formation of multienzyme complexes makes it possible to internally channel the product of one reaction to the next catalytic center without solvent exposure. Step 5: An intramolecular coupling reaction accompanied by a loss of water forms the five-membered imidazole ring. The carbonyl is O AIR synthetase C H ATP NH N7 N C8 ADP + Pi HC CH C5 NH P-ribose C CH2 P-ribose - H2O N N N9 C C4 N3 NH2 ring closure H Formylglycinamidine ribonucleotide 5-aminoimidazole ribonucleotide activated by phosphorylation and the phosphate is displaced by an amino group as shown on p. 5 (top) Step 6: Bicarbonate adds first to the exocyclic amino group and then is transferred to the neighboring carbon atom of the imidazole ring: ATP + O HO N N HC N O- CH HC CH N C P-ribose ADP + Pi - H2O NH2 8 carboxyl transfer HC O C 9N N 7 O 5 C 6 O- C 4 HN P-ribose carboxylation of the amino group O- P-ribose NH2 Carboxyaminoimidazole ribonucleotide 5-aminoimidazole ribonucleotide ATP + O HO H N Enzyme: AIR carboxylase HC CH O- H N ADP + Pi HC N CH C 8 carboxyl transfer HC H 7 N O 5 C 6 O O- C 9N C 4 - H Ocarboxylate is phosphorylated, Step 7:N The imidazole and the HN P-ribose P-ribose NH P-ribose NH phosphate is displaced by the amino group ofO- aspartate: 2 2 2 carboxylation of the amino group Carboxyaminoimidazole ribonucleotide 5-aminoimidazole ribonucleotide 5 ATP O N ADP + Pi HC O- C P-ribose - H2O H2N Carboxyaminoimidazole ribonucleotide CH CO2- CH CH2 NH2 CO2- 5-aminoimidazole-4-(N-succinylcarboxamide) ribonucleotide CH2 Asp N H C N P-ribose NH2 CO2- C C N O N HC CO2- Enzyme: SAICAR synthetase Step 8. Elimination of fumarate is catalyzed by lyase. The carbon skeleton of aspartate is lost as fumarate (only the amino group is contributed by aspartate). CO2CH fumarate HC HC N CO2- O N C N H C P-ribose CH N1 HC CH H NH2 O N CO2- N CO2- C NH2 C P-ribose NH2 5-aminoimidazole-4-carboxamide ribonucleotide 5-amidoimidazole-4-(N-succinylcarboxxamide) ribonucleotide Step 9: The second formyl group is added from N10formyltetrahydrofolate: O N HC N P-ribose 10 N -formylTHF C O N HC THF C NH2 C N NH2 P-ribose 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) NH2 C NH C2 C H O 5-formaminoimidazole-4-carboxamide ribonucleotide (FAICAR) 6 Enzyme: AICAR transformylase Step 10: Cyclization/dehydration produces inosinate: HC N 7 O N 8 NH2 C P-ribose NH 9 C N 4 P-ribose H C O 5 HC AMP cyclohydrolase C N 6 C 3 NH 1 N H2O CH 2 O Inosinate (IMP) 5-formaminoimidazole-4-carboxamide ribonucleotide (FAICAR) Formation of AMP and GMP from IMP IMP is a common precursor to both AMP and GMP. 1. Formation of adenylate (AMP): the C-6 carbonyl group of inosinate is replaced with the amino group from Asp. O- Adenylsuccinate synthetase O IMP dehydrogenase O O GDP + Pi H2O N NH N - H2O C ATP N N NN H P-ribose NN P-ribose P-ribose P-ribose H2N (IMP) (IMP) Asp = CH C CH2 C O- Xanthylate Adenylsuccinate (XMP) H H O O COO- AMP + PPi NH2 N N NH O- H2O O O Inosinate Inosinate CH HN NH NH N N N NADH + H+ N N fumarate CH NAD+ Asp H GMP synthetase GTP O -OOC C N N N H3N P-ribose P-ribose + lyaseH2O + Gln Glu N N Guanylate (GMP) Adenylate (AMP) O OH Note that GTP is used as a phosphate donor 2. Formation of guanylate (GMP). Inosinate(Inosine instead of ATP monophosphate) is first oxidized to xanthylate, and the C-2 carbonyl is then converted to an amino group. 7 NH2 Regulation of Purine Biosynthesis: AMP, GMP and IMP are feedback inhibitors of purine biosynthesis Purine biosynthesis is energetically expensive (6 moles ATP used per 1 mol IMP) Nucleotide concentrations are tightly regulated in a cell. Loss of regulation may lead to clinical disease (e.g. gout). 1. PRPP synthesis is inhibited by AMP, IMP, GMP (indicators of poor energy status) 2. The committed step (phosphoribosylamine formation) is inhibited by all purine mononucleotides. 3. AMP and GMP inhibit their own synthesis from IMP nucleotide Reciprocal substrate regulation balances the relative purine levels: AMP synthesis requires GTP as the energy source GMP synthesis requires ATP as the energy source Purine biosynthesis: 1. Purines are synthesized using both de novo and salvage pathways. 2. The purine ring is built on a ribose skeleton to make IMP, followed by branches to AMP and GMP. Amino acids serve as N 8 donors, while CO2 and C1 units of N10-formyl-THF are carbon donors. 3. The general mechanism involves ATP-mediated phosphorylation of carbonyl oxygen, followed by phosphate displacement by an amine. 4. Purine biosynthesis is tightly regulated via feedback inhibition. Formation of phosphoribosylamine is the committed step. The loss of regulation can lead to a clinical disease. 5. NAD+ is biosynthesized from nicotinate, ATP, and PRPP. Glutamine is used as an amino donor. REFERENCES: 1. ^ Rosemeyer, H. Chemistry & Biodiversity 2004, 1, 361. 2. ^ McGuigan, Hugh (1921). An Introduction To Chemical Pharmacology. P. Blakiston's Sons & Co. p. 283. Retrieved July 18, 2012. 3. ^ Fischer, E. Berichte der Deutschen Chemischen Gesellschaft 1899, 32, 2550. 4. ^ Scheele, V. Q. Examen Chemicum Calculi Urinari, Opuscula, 1776, 2, 73. 5. ^ http://www.dietaryfiberfood.com/purine-food.php 6. ^ Gout Diet: Limit High Purine Foods 7. ^ NEJM - Purine-Rich Foods, Dairy and Protein Intake, and the Risk of Gout in Men 8. ^ Callahan; Smith, K.E.; Cleaves, H.J.; Ruzica, J.; Stern, J.C.; Glavin, D.P.; House, C.H.; Dworkin, J.P. (11 August 2011). "Carbonaceous meteorites contain a wide range of extraterrestrial nucleobases". PNAS. doi:10.1073/pnas.1106493108. Retrieved 2011-0815. Unknown parameter |unused_data= ignored (help) 9. ^ Steigerwald, John (8 August 2011). "NASA Researchers: DNA Building Blocks Can Be Made in Space". NASA. Retrieved 2011-08-10. 10.^ Yamada, H.; Okamoto, T. (1972). "A One-step Synthesis of Purine Ring from Formamide".Chemical & Pharmaceutical Bulletin 20 (3): 623. doi:10.1248/cpb.20.623. 11.^ Saladino et al.; Crestini, Claudia; Ciciriello, Fabiana; Costanzo, Giovanna; Mauro, Ernesto (2006). "About a Formamide-Based Origin of Informational 9 Polymers: Syntheses of Nucleobases and Favourable Thermodynamic Niches for Early Polymers". Origins of Life and Evolution of Biospheres 36 (5–6): 523– 531. Bibcode:2006OLEB...36..523S.doi:10.1007/s11084-006-90532. PMID 17136429. 12.^ Sanchez, R. A.; Ferris, J. P.; Orgel, L. E. (1967). "Studies in prebiotic synthesis. II. Synthesis of purine precursors and amino acids from aqueous hydrogen cyanide". Journal of Molecular Biology 30 (2): 223–53. PMID 4297187. 13.^ Ferris, J. P.; Orgel, L. E. (1966). Journal of the American Chemical Society 88 (5): 1074.doi:10.1021/ja00957a050. 14.^ Ferris, J. P.; Kuder, J. E.; Catalano, O. W.; Kuder; Catalano (1969). "Photochemical Reactions and the Chemical Evolution of Purines and Nicotinamide Derivatives". Science 166 (3906): 765– 6. Bibcode:1969Sci...166..765F. doi:10.1126/science.166.3906.765.PMID 42418 47. 15.^ Oro, J.; Kamat, J. S.; Kamat (1961). "Amino-acid Synthesis from Hydrogen Cyanide under Possible Primitive Earth Conditions". Nature 190 (4774): 442– 3.Bibcode:1961Natur.190..442O. doi:10.1038/190442a0. PMID 13731262. 16.^ Houben-Weyl, Vol . E5, p. 1547[full citation needed] 17.^ Organic Syntheses Based on Name Reactions, Alfred Hassner, C. Stumer ISBN 008043259X2002 10