Supplemental information accompanying the Behavioral Ecology
advertisement

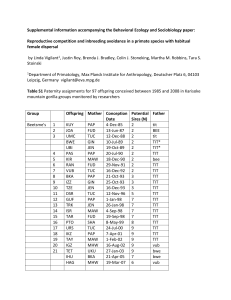
1 2 3 4 5 6 7 8 9 10 Supplemental information accompanying the Behavioral Ecology and Sociobiology paper: 11 12 13 Supplementary methods 14 Reproductive competition and inbreeding avoidance in a primate species with habitual female dispersal by Linda Vigilant1, Justin Roy, Brenda J. Bradley, Colin J. Stoneking, Martha M. Robbins, Tara S. Stoinski 1 Department of Primatology, Max Planck Institute for Anthropology, Deutscher Platz 6, 04103 Leipzig, Germany vigilant@eva.mpg.de Genetic analysis Most samples were collected using a two-step method of short-term storage in alcohol 15 followed by desiccation, but some earlier samples were simply desiccated using silica gel or 16 stored in RNAlater (storage details as described in (Nsubuga et al. 2004). We extracted DNA 17 using the QIAmp Stool kit (Qiagen). 18 For genotyping we used 13 of the 15 loci employed previously in a study of this 19 population (Bradley et al. 2005) and replaced the previously used D7s794 and D18s851 with 20 D1s2130, D5s1457, and D8s1106. We used a multiplexing protocol in which all primers were 21 included in an initial PCR followed by re-amplification at each single locus. Primer sequences 22 and PCR reaction conditions are as previously detailed for mountain gorillas (Arandjelovic et al. 23 2009). Dilutions of PCR products were combined in sets of three or four with ROX labeled 24 HD400 as an internal size standard and electrophoresed on an ABI Prism 3100 Genetic Analyzer 25 and analyzed using GeneMapper software v. 3.7 (Applied Biosystems). 26 Whenever possible we extracted DNA and produced genotypes from more than one 27 sample per individual in order to confirm attribution of the samples to particular individuals. We 28 performed extensive replication of PCRs according to the amount of DNA present in the extracts 29 as described in: (Arandjelovic et al. 2009) in order to guard against genotyping error, particularly 30 allelic dropout and false inference of a homozygous genotype. In practice, homozygous 31 genotypes were observed at least 3 times and often 6 or more times. In addition, we checked that 32 genotypes of known relatives (ie., mother-offspring) were genetically compatible, and confirmed 33 the sex of the individual using a PCR-based assay as previously described (Arandjelovic et al. 34 2009). 35 36 37 General Linear Mixed Models We used a binomial variance model and logit link function. The log odds ratio of the 38 inverse number of males was included as an offset term. The male ID, mother ID and offspring 39 ID were included as multilevel random effects. All predictors were transformed to have zero 40 mean and unit standard deviation. Predictors that represented timespans (age, tenure length) were 41 additionally square-root-transformed prior to this step. We checked for multicolinearity among 42 the predictors by computing the model matrix condition number; condition numbers greater than 43 30 were taken as evidence of potentially interfering multicolinearity (Belsey et al. 1980). The p 44 values were based on a likelihood ratio test, which avoids problems with p values based on a 45 normal approximation (Hauck and Donner 1977). The p values were determined from an initial 46 model fit using all relevant predictors, and adjusted for multiple comparisons using the Holm 47 procedure (Holm 1979). After fitting the initial full model, we removed predictors stepwise until 48 the Akaike information criterion was minimal (Sakamoto et al. 1986). We do not report p values 49 for the predictors in these reduced models, because they may be influenced by the model 50 reduction. In order to evaluate how well our best-fitting models actually explained the data, we 51 plotted receiver operating characteristic (ROC) curves, using the ROCR package in R (Sing et al. 52 2005). We used the area under the ROC curve as a convenient summary value (Bradley 1997). 53 54 Supplementary results 55 56 Genotyping and parentage assignment. The genotypes of 149 gorillas at 16 autosomal 57 microsatellite loci were 95% complete. There was an average of 4.63 alleles and an expected 58 heterozygosity of 0.55. The ability to determine paternity was high, with a combined non- 59 exclusion probability of 0.0017 given the genotype of the mother. One locus (D4s1627) was 60 found to not be in Hardy-Weinberg equilibrium after correction for multiple testing, but since the 61 data consists of a structured population containing many relatives this result is not unusual and is 62 not expected to affect the paternity determinations. The paternity assigments produced using 63 CERVUS had high (90%) likelihood and in all cases the assigned father had no genotypic 64 ‘mismatches’ to the offspring and other candidate males were excluded by one or more 65 mismatches. 66 Of the 47 paternities reported previously (Bradley et al. 2005), we reassigned MNZ, 67 formerly attributed to ZIZ, to PAB. The prior assignment was based on less complete data and 68 we now find that PAB is compatible at 16 loci with MNZ and the mother and hence was 69 assigned as the father by CERVUS with high likelihood while ZIZ mismatches at one of 16 loci. 70 All other previous assignments were confirmed, and we amend the previous unspecific paternity 71 assignment of TEG from ‘NotShinda’ to NTA. Three offspring (RIB, RAM, RUS) were 72 conceived in groups containing a single silverback male and in each case he was the sire. One 73 additional offspring (KEZ) was born in a group containing one silverback and one juvenile male, 74 and the silverback was the sire. The other 93 offspring assigned were conceived in groups 75 containing multiple silverback and potentially reproductive ‘blackback’ males. (Supplementary 76 Information Table S1). 77 With regard to the shares of paternity accruing to the dominants (Table 1 main text), for 78 one male (BEE) we assess only three paternities from the final four years of his tenure, and 79 similarly analyze paternities produced in the final seven years of the approximately eight year 80 tenure of ZIZ. For the other males, we analyzed offspring produced during the entire 13 year 81 tenures of SHI and TIT, respectively and during 11 years of the ongoing tenure of CAN. 82 83 Cases of female dispersal. In the BEE group, two nulliparous females (UMC, PAS) transferred 84 to different groups in 1999 at the ages of 9.4 and 8.5. IZZ did not produce offspring in her natal 85 group (BEE) but only after dispersing at the relatively late age of 12.8. In Group 5, the daughters 86 of the dominant ZIZ did not reach typical dispersal age before the death of the dominant and 87 subsequent formation of PAB and SHI groups. Two females (UKU, MHW) were fathered by 88 PAB in 1993 during a period for which it is unknown whether PAB or CAN was the dominant 89 male, and both dispersed from their natal group, which by then featured CAN as dominant male, 90 by the ages of 7.7 and 7.3. Both daughters of SHI (KAN, KRD) that were of dispersal age 91 stayed and conceived in the natal group. 92 93 Supplementary References 94 95 Arandjelovic M, Guschanski K, Schubert G, Harris TR, Thalmann O, Siedel H, Vigilant L (2009) Two- 96 step multiplex polymerase chain reaction improves the speed and accuracy of genotyping using 97 DNA from noninvasive and museum samples. Mol Ecol Resour 9:28-36 98 99 100 101 102 Belsey DA, Kuh E, Welsch RE (1980) Regression Diagnostics: Identifying Influential Data and Sources of Collinearity. John Wiley & Sons, Hoboken Bradley AP (1997) The use of the area under the roc curve in the evaluation of machine learning algorithms. Pattern Recogn 30:1145-1159 Bradley BJ, Robbins MM, Williamson EA, Steklis HD, Steklis NG, Eckhardt N, Boesch C, Vigilant L 103 (2005) Mountain gorilla tug-of-war: Silverbacks have limited control over reproduction in 104 multimale groups. Proc Nat Acad Sci USA 102:9418-9423 105 106 Hauck WW, Donner A (1977) Walds test as applied to hypotheses in logit analysis. J Am Stat Assoc 72:851-853 107 Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6:65-70 108 Nsubuga AM, Robbins MM, Roeder AD, Morin PA, Boesch C, Vigilant L (2004) Factors affecting the 109 amount of genomic DNA extracted from ape faeces and the identification of an improved sample 110 storage method. Mol Ecol13:2089-2094 111 Sakamoto Y, Ishiguro M, Kitagawa G (1986) Akaike information criterion statistics. D. Reidel, Dordrecht 112 Sing T, Sander O, Beerenwinkel N, Lengauer T (2005) ROCR: visualizing classifier performance in R. 113 114 Bioinf 21:3940-3941.