TableS1: List of variants confirmed to be true by Sanger sequencing
advertisement
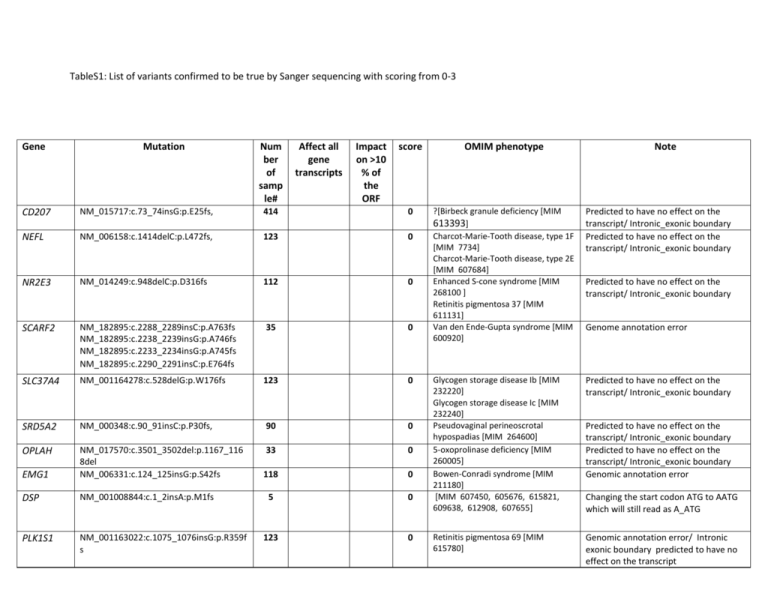
TableS1: List of variants confirmed to be true by Sanger sequencing with scoring from 0-3 Gene Mutation Num ber of samp le# Affect all gene transcripts Impact on >10 % of the ORF score OMIM phenotype ?[Birbeck granule deficiency [MIM 613393] Charcot-Marie-Tooth disease, type 1F [MIM 7734] Charcot-Marie-Tooth disease, type 2E [MIM 607684] Enhanced S-cone syndrome [MIM 268100 ] Retinitis pigmentosa 37 [MIM 611131] Van den Ende-Gupta syndrome [MIM 600920] Predicted to have no effect on the transcript/ Intronic_exonic boundary Predicted to have no effect on the transcript/ Intronic_exonic boundary Glycogen storage disease Ib [MIM 232220] Glycogen storage disease Ic [MIM 232240] Pseudovaginal perineoscrotal hypospadias [MIM 264600] 5-oxoprolinase deficiency [MIM 260005] Bowen-Conradi syndrome [MIM 211180] [MIM 607450, 605676, 615821, 609638, 612908, 607655] Predicted to have no effect on the transcript/ Intronic_exonic boundary Retinitis pigmentosa 69 [MIM 615780] Genomic annotation error/ Intronic exonic boundary predicted to have no effect on the transcript CD207 NM_015717:c.73_74insG:p.E25fs, 414 0 NEFL NM_006158:c.1414delC:p.L472fs, 123 0 NR2E3 NM_014249:c.948delC:p.D316fs 112 0 SCARF2 NM_182895:c.2288_2289insC:p.A763fs NM_182895:c.2238_2239insG:p.A746fs NM_182895:c.2233_2234insG:p.A745fs NM_182895:c.2290_2291insC:p.E764fs 35 0 SLC37A4 NM_001164278:c.528delG:p.W176fs 123 0 SRD5A2 NM_000348:c.90_91insC:p.P30fs, 90 0 OPLAH 33 0 EMG1 NM_017570:c.3501_3502del:p.1167_116 8del NM_006331:c.124_125insG:p.S42fs 118 0 DSP NM_001008844:c.1_2insA:p.M1fs 5 0 PLK1S1 NM_001163022:c.1075_1076insG:p.R359f s 123 0 Note Predicted to have no effect on the transcript/ Intronic_exonic boundary Genome annotation error Predicted to have no effect on the transcript/ Intronic_exonic boundary Predicted to have no effect on the transcript/ Intronic_exonic boundary Genomic annotation error Changing the start codon ATG to AATG which will still read as A_ATG Combined hyperlipidemia, familial [MIM 144250] Lipoprotein lipase deficiency [MIM 238600] Deafness, autosomal dominant 11 [MIM [601317] Deafness, autosomal recessive 2 [MIM 600060] Usher syndrome, type 1B [MIM 276900] Aicardi-Goutieres syndrome 2 [MIM 610181] LPL NM_000237.2:c.1421C>G ; p.Ser474* 7 no no 1 MYO7A NM_001127179.2:c.3515_3536del; p.Gly1172Glufs*34 68 no no 1 RNASEH2B NM_024570.3: c.925dup; p.Ile309Asnfs*8 2 no no 1 TREH NM_007180.2:c.1706dupC : p.Ala570Glyfs*35 122 no no 1 Trehalase deficiency [MIM 612119] ABHD12 NM_001042472.2:c.1189C>T; p.Gln397* 1 no no 1 No phenotype associated with the respective OMIM phenotype. SETBP1 NM_001130110.1:c.681_682insTCTT ;p.Thr228Serfs*8 123 no no 1 ANTXR2 NM_058172.5:c.1450C>T: p.Arg484* 1 no no 1 H6PD NM_001282587.1:c.-7-0delCCCAGGCA 7 no yes 2 LFNG NM_001166355.1: c.163_166dup 16 no yes 2 Polyneuropathy, hearing loss, ataxia, retinitis pigmentosa, and cataract [MIM 612674] Mental retardation, autosomal dominant 29 [MIM 616078 ]/ Schinzel-Giedion midface retraction syndrome [MIM 269150] Hyaline fibromatosis syndrome [MIM 228600 ] Cortisone reductase deficiency 1 [MIM 604931] ?Spondylocostal dysostosis 3, autosomal recessive [MIM 609813] Amyloidosis, primary localized cutaneous, 1/ Autosomal dominant Ichthyosis, congenital, autosomal recessive 10 [ 615024] Mitochondrial DNA depletion syndrome 8A and B [MIM 612075] Progressive external ophthalmoplegia with mitochondrial DNA deletions, No phenotype associated with the respective OMIM phenotype No phenotype associated with the respective OMIM phenotype No phenotype associated with the respective OMIM phenotype p.Glu56Glyfs*2 OSMR NM_003999.2:c.1207C>T; p.Arg403Ter 1 no yes 2 PNPLA1 NM_001145716.2:c.1206T>A; p.Tyr402* 1 yes no 2 RRM2B NM_001172477.1:c.211dup ; p.Arg71Profs*28 1 no yes 2 No phenotype associated with the respective OMIM phenotype Phenotype could not be assessed _early death autosomal dominant, 5 [MIM 613077] TGIF1 NM_170695.3:c.246delT; p.Pro83Leufs*51 41 no yes 2 Holoprosencephaly-4 [MIM 602630] LTBP4 NM_001042544.1:c.3236dupG; p.Tyr1080Leufs*17 120 no yes 2 Cutis laxa, autosomal recessive, type IC [MIM 613177] TMPRSS15 NM_002772.2:c.2808_2809insATCA; p.Ser937Ilefs*4 1 yes no 2 Enteropeptidase deficiency [MIM 226200] Phenotype could not be assessed _early death AMPD1 NM_000036.2:c.133C>T ;p.Gln45* 2 no yes 2 AMT NM_001164711.1:c.814dup; p.Ala272Glyfs*22 NM_001081563.2:c.1691dup; p.Ala565Glyfs*28 1 no yes 2 1 no yes 2 Myopathy due to myoadenylate deaminase deficiency [MIM 615511] Glycine encephalopathy [MIM 605899] Myotonic dystrophy 1 [MIM 160900] CDH15 NM_004933.2: c.2088_2089del; p.His697Profs*20 1 no yes 2 Mental retardation, autosomal dominant 3 [MIM 612580] No phenotype associated with the respective OMIM phenotype Phenotype could not be assessed _early death Reported mutations in this gene only as repeat variation/Autosomal dominant No phenotype associated with the respective OMIM phenotype No phenotype associated with the respective OMIM phenotype CCDC39 NM_181426.1:c.2660dupT; p.Ser888Ilefs*6 1 yes no 2 Ciliary dyskinesia, primary, 14 [MIM 613807] No phenotype associated with the respective OMIM phenotype COQ6 NM_182480.2:c.41G>A; p.Trp14* 1 no yes 2 CC2D2A NM_020785.2: c.262C>T; p.Arg88* 2 No yes 2 No phenotype associated with the respective OMIM phenotype No phenotype associated with the respective OMIM phenotype FLG NM_002016.1:c.12181G>T; p.Glu4061* 1 yes no 2 ALG9 NM_024740.2:c.61del;p.Ala21Profs*55 75 no yes 2 PRSS12 NM_003619.3:c.2323C>T : p.Arg775* 1 yes yes 3 Coenzyme Q10 deficiency, primary, 6 [MIM 614650] COACH syndrome [MIM 216360] Joubert syndrome 9 [MIM 612285] Meckel syndrome 6 [MIM 612284] Ichthyosis vulgaris [MIM 146700] Dermatitis, atopic, susceptibility to, 2 [MIM 605803] Congenital disorder of glycosylation, type Il [MIM 608776 ] Mental retardation, autosomal recessive 1 [MIM 249500] DMPK Phenotype could not be assessed _early death See Table 1 and text Erythrokeratodermia variabilis with erythema gyratum repens/ Deafness [MIM 133200] Spermatogenic failure 14 [MIM 615842] Ichthyosis, congenital [MIM 613943] See Table 1 and text 3 Poikiloderma, hereditary fibrosing, with tendon contractures, myopathy, and pulmonary fibrosis [MIM 615704] See Table 1 and text yes 3 Spinocerebellar ataxia 23 [MIM 610245] See Table 1 and text yes yes 3 Familial cold autoinflammatory syndrome 2 [MIM 611762] See Table 1 and text 1 yes yes 3 Hyperglycinuria [MIM 138500] See Table 1 and text 40 yes yes 3 10 yes yes 3 ABCG5 NM_022436.2:c.1336C>T: p.Arg446* 1 yes yes 3 Hypomagnesemia 3, renal [MIM 248250] Night blindness, congenital stationary (complete), 1C, [MIM 613216] Sitosterolemia [MIM 210250 See Table 1 and text TRPM1 NM_006580.3:c.166delG; p.Ala56Leufs*16 NM_001252020.1:c.4240G>T; p.Glu1414* GJB4 NM_153212.2: c.155_158del; p.Val52Alafs*55 1 yes yes 3 ZMYND15 NM_001267822.1: c.1024dupC; p.Leu342Profs*4 NM_001102469.1:c.302delG; p.Gly101Glufs*7 1 yes yes 3 3 yes yes 3 FAM111B NM_198947.3:c.394delT: p.Tyr132Metfs*6 2 yes yes PDYN NM_001190899.2:c.34delC; p.Leu12Serfs*20 2 yes NLRP12 NM_144687.3: c.1854C>G; p.Tyr618* 1 SLC36A2 NM_181776.2:c.1010G>A; p.Trp337* CLDN16 LIPN See Table 1 and text See Table 1 and text See Table 1 and text See Table 1 and text Score 3: Variants affect all gene transcripts and >10% of the ORF. Score 2: Variant either affect >10% of the ORF or affect all gene transcripts. Score 1: variants affect the last 10% of the ORF and only in a subset of the gene’s transcripts. Score 0: Variants with annotation error or predicted to be non LOF. # No of samples in which this variant confirmed by Sanger sequencing to present in homozygous state.