Title: Functional and feeding niche differences between
advertisement

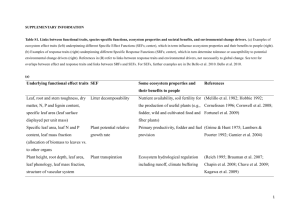
Supporting information Appendix S1. Grasshopper and plant functional traits 3 IS La/Li Ee 2 1 1/Ri Cb 56 % -4 Cd 0 -3 CN -2 -1 0 Pp 1 A 2 BS 3 Pg -2 39 % -1 Ci Fig. S1A. Co-variation between grasshopper species traits in a principal component analysis (PCA). Species are represented in black and traits in grey. The two orthogonal axes explain respectively 56% and 39% of the total variance..Grasshopper species abbreviations are: Cb: C. biguttulus, Cd: C. dorsatus, Ci: C. italicus, Ee: E. elegantulus, Pg: P. giornae, Pp: P. parallelus. See the main text for grasshopper trait abbreviations. Method S1. Plant functional traits Plant functional traits were extracted from a global survey conducted in the LTER (“Zone atelier Plaine et Val de Sèvre”) study site in 2012 on 25 grasslands representative of the grassland diversity in the studied region. Traits were measured using standardised protocols (Cornelissen et al. 2003). Within each of the 25 grasslands, we selected plant species representing more than 80% of the total biomass. All leaf trait measurements were carried out on 8 individuals per species. 84 species and 1802 individuals were sampled. In the present paper, we selected seven leaf traits related to (1) plant chemical properties: leaf nitrogen content (LNC), carbon/nitrogen ratio (C:N) and specific leaf area (SLA), (2) plant biomechanical properties: leaf dry matter content (LDMC), leaf thickness (LT) and leaf carbon content (LCC) and (3) architectural properties: leaf area (LA), and perimeter / leaf length ratio (P:LL). 25% 3 LDMC Er Pa Fr Be Fa Bh C:N -2 Pr Pl Am Lv PmCsp Pe Sj Esp 2 RfAe Dg 1Pp Pt Gv 0 -1 LT 0 Dc Ra Sp VoCj -1 LCC Tc Pr Tp P:LLVs Ms LNCLc SLA Ca Tr Lp 1 2 Gr GmTo Or 48% 3 Ph -2 Ra -3 Fig. S1B. Co-variation between plant species traits in a principal component analysis (PCA). Plant species are represented in three different colours (red for legumes, blue for other forbs and green for grasses). Species functional traits are represented in black. The two orthogonal axes explain respectively 48% and 25% of the variance. Trait abbreviations are: LDMC: leaf dry matter content, LCC: leaf carbon content, P:LL: perimeter / leaf length ratio, LNC: leaf nitrogen content, LA: leaf area , LT: leaf thickness, C:N: carbon / nitrogen ratio. Species abbreviations are: Ae: Arrhenatherum eliatus, Am: Achillea millefolium, Be: Bromus erectus, Bh: Bromus hordeaceus, Cj: Centaurea jacea, Ca: Convolvulus arvensis, Csp: Crepis sp, Dg: Dactylis glomerata, Dc: Daucus carota, Er: Elitrigia repens, Esp: Erigeron sp, Fa: Festuca arundinacea, Fr: Festuca rubra, Ga: Galium aparine, Gm: Galium mollugo, Gr: Geranium rotundifolium, Gv: Galium verum, Lc: Lotus corniculatus, Lp: Lolium perenne, Lv: Leucanthemun vulgare, Ms: Malva sylvestris, Or: Ononis repens, Pa: Poa angustifolia, Pe: Picris echoides, Ph: Picris hieracioides, Pl: Plantago lanceolata, Pm: Plantago major, Pp: Poa pratensis, Pr: Potentilla reptans, Pt: Poa trivialis, Pv: Prunella vulgaris, Ra: Ranunculus acris, Re: Rumex acetosela, Rf: Rubus fruticosus, Sj: Senecio jacobaea, Sp : Salvia pratensis, Tc: Trifolium campestre, To : Tarxacum officinalis, Tp: Trifolium pretense, Tr: Trifolium repens, Vo: Verbena officinalis, Vs: Vicia sativa