LAB 2: Connecting Population Growth and Biological Evolution
advertisement

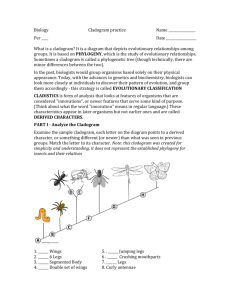
NAME ______________________________________ DATE ____________ PER _________ Structural and Molecular Homologies Objectives: Given some groups of organisms and their distinguishing characteristics, you will construct a cladogram based on the homologous structures. Then you will interpret and analyze that cladogram in terms of how it shows common ancestry and degrees of evolutionary relationship. Then you will analyze the gene for cytochrome C in several different organisms and see and how it relates to the cladogram you made. Part 1: Structural Homologies Procedure: 1. Determine which of the characteristics each animal has. In the Data Table provided (on your Cladogram Worksheet), place an "x" in the box if the animal has the characteristic. Explanations of Characteristics: set #1: Dorsal nerve cord (running along the back or "dorsal" body surface) Notochord (a flexible but supporting cartilage-like rod running along the back or "dorsal" surface) set #2: Paired appendages (legs, arms, wings, fins, flippers, antennae) Vertebral ("backbone") set #3: Paired legs set #4: Amnion (a membrane that holds in the amniotic fluid surrounding the embryo; may or may not be inside an egg shell) set #5: Mammary glands (milk-secreting glands that nourish the young) set #6: Placenta (structure attached to inside of uterus of mother, and joined to the embryo by the umbilical cord; provides nourishment and oxygen to the embryo) set #7: Canine teeth short (same length as other teeth) Foramen magnum forward (spinal cord opening, located forward, under skull) 2. Using the chart of the groupings just completed, draw a cladogram on your sheet to illustrate the ancestry of these animals. The diagram should reflect shared characteristics as time proceeds. Structural Analysis Questions 1. According to the structural homologies, which organism is most related to the human? 2. According to the structural homologies, which organism is least related to the human? 3. What are the outgroups to the paired legs? 4. How many derived characters do the kangaroo and the monkey share? Part 2: Molecular Homologies Procedure: 3. Next, find the human, rhesus monkey, kangaroo, snapping turtle, bullfrog, and tuna on the "Amino Acid Sequences in Cytochrome-C Proteins from 20 Different Species" chart provided and underline their names. 4. Compare the human amino acid sequence with each of these five animals by counting the number of times an amino acid in that animal’s cytochrome c is different from the amino acid in that same position of the human sequence. For example, the number of differences between human and dog=10. Write that information below: Number of amino acid differences between human and the • Rhesus monkey= • Kangaroo= • Snapping turtle= • Bullfrog= • Tuna= 5. Record the total number of amino acid differences between humans and each animal shown below. Write your answer in the hexagon below the arrow pointing to the name of that animal. Molecular Analysis Questions 1. Does the data from the amino acid sequence generally agree with the anatomical data that was used to make the cladogram? 2. Do organisms with fewer shared anatomical traits also have more amino acid differences? 3. Based on the molecular data, how does the "human-monkey" relationship compare to the "duckchicken" relationship (which shows three amino acid differences)? 4. If the molecular data, the structural similarities, and the fossil record all support the same pattern of relationships, can we be fairly confident that the pattern is accurate? Why or why not? 5. Chickens and turkeys are both birds and have the same sequence of amino acids in their cytochrome-c protein. Explain how two species can have identical cytochrome-c and still be different species. 6. Neurospora (bread mold) and Saccharomycetes (bakers yeast) are both fungi. Chickens and turkeys are both birds. What can you say about the inferred evolutionary relationships between the two birds compared to the relationship between the two fungi? Explain your reasoning.