emi12685-sup-0001-si
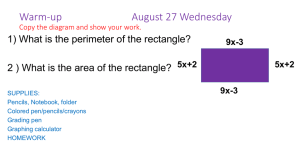
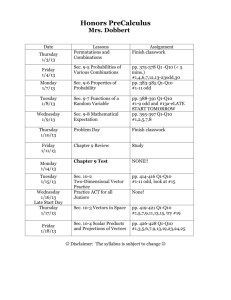
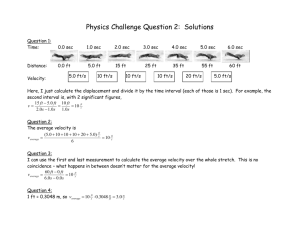
advertisement

1 Supporting information 2 Table S1. PCR Results from mussel hemolymph spiking experiments showing number of 3 samples positive for T. gondii / number of replicates for (A) screening assays using primers 4 targeting multi-copy genes including ITS1, B1, and a 529 bp repetitive element (RE); and (B) 5 genotyping assays using primers targeting single-copy genes including SAG1, SAG3, and 6 GRA6. The RE primer set yielded results that were identical to ITS1, and thus was not further 7 included in multiplexed assays or for testing field-collected mussels for T. gondii. The B1 primer 8 sets were retained for testing field-collected mussels due to the dual application of this locus for 9 further genotyping and molecular characterization. 10 (A) Assay type Spiking Multiplexed Simplex concentration oocysts / mL ITS1 B1 ITS1 B1 RE 0 0/3 0/3 0/3 0/3 0/3 1 0/3 0/3 0/3 0/3 0/3 5 1/3 0/3 1/3 1/3 1/3 10 1/3 0/3 1/3 0/3 1/3 50 3/3 2/3 3/3 3/3 3/3 100 3/3 2/3 3/3 3/3 3/3 1000 3/3 3/3 3/3 3/3 3/3 hemolymph 11 1 12 (B) Assay type Spiking Multiplexed Simplex concentration oocysts / mL SAG1 SAG3 GRA6 SAG1 SAG3 GRA6 0 0/3 0/3 0/3 0/3 0/3 0/3 1 0/3 0/3 0/3 0/3 0/3 0/3 5 0/3 0/3 0/3 0/3 0/3 0/3 10 0/3 0/3 0/3 0/3 0/3 0/3 50 1/3 1/3 0/3 0/3 1/3 0/3 100 1/3 2/3 1/3 1/3 1/3 1/3 1000 3/3 3/3 3/3 3/3 3/3 3/3 hemolymph 13 14 2 15 Table S2. Primer sequences and thermocycler conditions used in polymerase chain reaction assays for detection of T. gondii DNA in 16 mussel hemolymph. Loci used for screening assays (ITS1, B1, and the 529 bp repetitive element (RE)) were tested individually as 17 simplex assays as well as multiplexed, and loci used only for genotyping (SAG1, SAG3, and GRA6) were tested individually or 18 multiplexed together. Note that the B1 locus can be used both for screening and genotyping, as it is a multi-copy gene containing 19 single nucleotide polymorphisms. In multiplexed assays, primers for all loci were combined in the external reaction, and then primers 20 for each locus were used separately for the internal reaction, following the approach described by Su et al. (2010). Locus Purpose Thermocycler conditions Primer sequences a Restriction Reference Enzyme b ITS1 Screening Denaturation 3 min at 94C; 35 External forward: cycles: 40 sec at 95C, 40 sec TACCGATTGAGTGTTCCGGTG at 58C (external) or 59C External reverse: (internal), 90 sec at 72C; Post PCR extension 4 min at 72C, hold at 4C GCAATTCACATTGCGTTTCGC Internal forward: CGTAACAAGGTTTCCGTAGG Internal reverse: TTCATCGTTGCGCGAGCCAAG 3 NA (Rejmanek et al., 2009) B1 Screening & External forward: genotyping TGTTCTGTCCTATCGCAACG PmlI, XhoI External reverse: (Grigg and Boothroyd, 2001) ACGGATGCAGTTCCTTTCTG Internal forward: TCTTCCCAGACGTGGATTTC Internal reverse: CTCGACAATACGCTGCTTGA RE Screening Denaturation: 7 min at 94C; External forward: 35 cycles: 60 sec at 94C, 60 CGCTGCAGGGAGGAAGACGAAA sec at 55C, 60 sec at 72C; GTTG Post PCR extension: 10 min at External reverse: 72C NA (Homan al., 2000) (Shapiro CGCTGCAGACACAGTGCATCTGG ATT Internal forward: AGAAGGGACAGAAGTCGAAG 4 et al., 2010) et Internal reverse: CTCCACTCTTCAATTCTCTCC SAG1 Genotyping Denaturation 3 min at 94C External forward: (internal) or 95C (external); GTTCTAACCACGCACCCTGAG 35 cycles: 40 sec at 95C External Reverse: (external) or 30 sec at 94C (internal), 40 sec at 58C (external) or 30 sec at 60C Hae II, Sau96 I (Grigg et al., 2001) GTGGTTCTCCGTCGGTGTGAG Internal forward: CAATGTGCACCTGTAGGAAGC Internal reverse: (internal), 90 sec at 72C TTATCTGGGCAGGTGACAAC (external) or 45 sec at 72C SAG3 Genotyping (internal); Post PCR extension External forward: AlwNI, (Grigg et al., CAACTCTCACCATTCCACCC NciI 2001) 4 min (external) or 10 min External Reverse: (internal) at 72C, hold at 4C GCGCGTTGTTAGACAAGACA Internal forward: TCTTGTCGGGTGTTCACTCA 5 Internal reverse: CACAAGGAGACCGAGAAGGA GRA6 Genotyping External forward: MseI ATTTGTGTTTCCGAGCAGGT (Fazaeli et al., 2000) External Reverse: GCACCTTCGCTTGTGGTT Internal forward: TTTCCGAGCAGGTGACCT Internal reverse: TCGCCGAAGAGTTGACATAG All primer sequences are listed in 5’to 3’ orientation. All PCR reactions were carried out in 50 l reaction volumes with 5 l of DNA 21 a 22 template used in external reactions and 2 l DNA amplicon from the external reaction used in internal reactions. 23 b 24 SAG1, SAG3, and GRA6). 25 NA = Not applicable Restriction enzymes used for genotyping with restriction fragment length polymorphism (RFLP) assays for genotyping loci (B1, 26 27 6 28 References 29 30 31 Fazaeli, A., Carter, P.E., Darde, M.L., and Pennington, T.H. (2000) Molecular typing of Toxoplasma gondii strains by GRA6 gene sequence analysis. Int J Parasit 30: 637-642. 32 Grigg, M.E., and Boothroyd, J.C. (2001) Rapid identification of virulent type I strains of the protozoan pathogen Toxoplasma 33 gondii by PCR-restriction fragment length polymorphism analysis at the B1 gene. J Clin Microbiol 39: 398-400. 34 35 Grigg, M.E., Ganatra, J., Boothroyd, J.C., and Margolis, T.P. (2001) Unusual abundance of atypical strains associated with human ocular toxoplasmosis. J Infect Dis 184: 633-639. 36 Homan, W.L., Vercammen, M., De Braekeleer, J., and Verschueren, H. (2000) Identification of a 200- to 300-fold repetitive 529 37 bp DNA fragment in Toxoplasma gondii, and its use for diagnostic and quantitative PCR. Int J Parasitol 30: 69-75. 38 Rejmanek, D., Vanwormer, E., Miller, M.A., Mazet, J.A.K., Nichelason, A.E., Melli, A.C. et al. (2009) Prevalence and risk factors 39 associated with Sarcocystis neurona infections in opossums (Didelphis virginiana) from central California. Vet Parasitol 40 166: 8-14. 41 42 Shapiro, K., Mazet, J.A.K., Schriewer, A., Wuertz, S., Fritz, H., Miller, W.A. et al. (2010) Detection of Toxoplasma gondii oocysts and surrogate microspheres in water using ultrafiltration and capsule filtration. Water Res 44: 893-903. 43 44 7