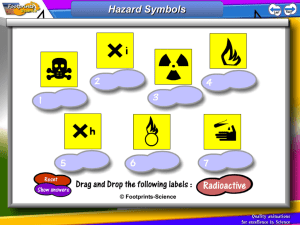
Full text - FNWI (Science) Education Service Centre
advertisement

Toxic compounds in saliva Development and validation of a microLC-QTOF screening method Master Thesis Thijs Meijer August 2015 University of Amsterdam Chemistry – Analytical Sciences MSc Chemistry Analytical Sciences Master Thesis Toxic compounds in saliva Development and validation of a microLC-QTOF screening method by Thijs Meijer August 2015 Supervisor: Dr. W.Th. Kok Daily Supervisor: Drs. Ing. M.H. Blokland ii Abstract Forensic veterinary toxicology is a growing field of interest. In The Netherlands only a veterinarian is allowed to take blood samples. This is a problem, since veterinarians are not always available when an acute intoxication takes place. Therefore, there is a need for toxicological analysis of alternative matrices. Saliva can be an interesting and animal friendly alternative. It can be sampled relatively easy without a veterinarian and it is a “mirror” of blood since it has comparable metabolism characteristics. Acute intoxication of animals can take place by intake of all kinds of toxic compounds. Three main compound groups were included in this study; pesticides, natural toxins and veterinary drugs. These compounds can be toxic for animals due to accidentally intake during feeding or negligence of the owner of the animal. They can also be administered on purpose like poisoning or overdosing. The aim of this research project was to develop a multi-residue method for the analysis of animal saliva using Liquid Chromatography – Mass Spectrometry (LC-MS). The development had to cover the entire procedure; saliva sampling, samples pre-treatment, LC-MS analysis and data interpretation. The method was validated according an in-house validation protocol based on official international legislation. As model animal saliva, calf saliva has been used, because the availability of calf saliva for this research was significantly higher compared to that of other animals. A sampling method was developed with two types of sampling devices, one for small volume and one for larger volume saliva sampling. The main reason for sample pre-treatment is to get rid of proteins which are present in the saliva. To achieve this a protein precipitation method using acetonitrile was developed and tested. A microLC-microfluidics-QTOF-MS method was developed for the identification of the compounds in saliva. With the microLC-microfluidics technique a relatively high sensitivity can be obtained using small sample injection volume and low flow rates. The microfluidics system consists of a Waters IonKey system, which includes an analytical column in a chip device (iKey), using ESI as an ionization technique. The IonKey source is placed in front of a QTOF MS. The acquisition method used is a so called MSe method where a full scan and a fragmentation scan are acquired alternately. In this way next to full scan data there is also fragmentation scan data available which can be used for more reliable identification of the compounds. A targeted library was developed using UNIFI, new MS software from Waters Corporation. The library includes retention times, monoisotopic masses and if present fragment ions of the toxic compounds which were included in the research project. The developed method was validated according international legislation (2002/657/EC) to test the characteristics of the method. For a screening method the limit of detection (CCß), selectivity and ruggedness of the method and stability of the compounds and sample extracts needs to be validated. The validation was performed on three days with 20 different blank saliva samples. The samples were spikes at two levels, 0.1 and 1 mg/liter. These corresponds with the CCß levels of the validated compounds. The validation results showed that with the method 203 compounds can be detected in saliva with a CCß of 0.1 or 1 mg/l. The method was not selective for twelve compounds, interfering peaks were found in the chromatograms. The method was rugged for two tested variations on the sample pre-treatment and sample extracts were proven to be stable for one week in the freezer. Within this research project a screening method for more than 200 toxic compounds was successfully developed. The method consists of a sampling procedure, sample pre-treatment and analyzing method using microLC-microfluidics-QTOF-MS. Additionally, the method was validated according European legislation. iii Contents Abstract .......................................................................................................................................... iii Contents .......................................................................................................................................... iv List of abbreviations ........................................................................................................................... v 1 Introduction .............................................................................................................................. 1 2 Background information .............................................................................................................. 4 3 2.1 Saliva ............................................................................................................................... 4 2.2 Transportation of toxic compounds in saliva ........................................................................... 5 2.3 Toxic compounds ................................................................................................................ 6 2.4 Liquid chromatography........................................................................................................ 6 2.5 Mass spectrometry ........................................................................................................... 10 2.6 Data handling .................................................................................................................. 11 2.7 Validation ........................................................................................................................ 12 Materials and Methods .............................................................................................................. 14 3.1 Materials ......................................................................................................................... 14 3.2 Methods .......................................................................................................................... 17 3.3 Validation .............................................................................................................................. 19 4 Results and discussion .............................................................................................................. 19 4.1 Sampling ......................................................................................................................... 21 4.2 Sample pre-treatment ....................................................................................................... 22 4.3 LC-MS analysis ................................................................................................................. 24 4.4 Validation ........................................................................................................................ 32 5 Conclusion .............................................................................................................................. 34 6 Recommendations and future perspectives .................................................................................. 35 7 References .............................................................................................................................. 36 Appendix I: Camera view of IonKey source interior .............................................................................. 37 Appendix II: TOF-MS calibration file ................................................................................................... 38 Appendix III: Compound list with RT and monoisotopic mass ................................................................ 39 Appendix III (continued): Compound list with RT and monoisotopic mass ............................................... 40 Appendix III (continued): Compound list with RT and monoisotopic mass ............................................... 41 Appendix IV: Processing method (partly) ............................................................................................ 42 Appendix V: Experimental scheme of validation day 3 .......................................................................... 43 Appendix VII: Validated compound with CCß values ............................................................................. 46 Appendix VII (continued): Validated compound with CCß values ............................................................ 50 Appendix VII (continued): Validated compound with CCß values ............................................................ 51 Appendix VII: Validated compound with CCß values ............................................................................. 46 Appendix VII (continued): Validated compound with CCß values ............................................................ 47 Appendix VII (continued): Validated compound with CCß values ............................................................ 48 Appendix VIII: Final sample pre-treatment method .............................................................................. 52 Appendix IX: Results of validation day 3 ............................................................................................. 53 iv List of abbreviations MeOH: methanol ACN: acetonitrile DMSO: dimethyl sulfoxide FA: formic acid LC: liquid chromatography MS: mass spectrometry TOF: time of flight UHPLC: ultra high performance (pressure) liquid chromatography IS: internal standard TIC: total ion chromatogram XIC: extracted ion chromatogram LOD: limit of detection LD50: median lethal dose GC: gas chromatography TLC: thin layer chromatography HPLC: high pressure (performance) chromatography SOP: standard operating procedure v 1 Introduction Forensic veterinary toxicology is a growing field of interest. It is a discipline concerned with the study of toxic substances or poisons in the veterinarian field. Reasons for this increased interest are growing commercial interests of livestock and the way animal owners are committed to their animals nowadays (1, 2). Chemical analyses of (biological) samples is an important aspect of veterinary toxicology for which purpose blood or serum samples traditionally are the main matrices of investigation. In The Netherlands only a veterinarian is allowed to take blood samples. This is a problem, since veterinarians are not always available when an acute intoxication takes place. Therefore, there is a need for toxicological analysis of alternative matrices. Matrices such as urine, hair, meat, organs, stomach content and eyes are already used for the detection of forbidden or toxic substances (3) (see table 1). However, all these matrices have disadvantages such as sampling time, contamination risk, the invasive nature of the sampling procedure for the animal or an extensive sample pre-treatment procedure. Saliva can be an interesting and animal friendly alternative to these matrices. It can be sampled relatively easy without a veterinarian and it is a “mirror” of blood since it has comparable metabolism characteristics (4, 5). So, using saliva instead of blood makes it possible to undertake action in a more early state of the intoxication, because no veterinarian is involved. It can be done relatively simple and sample pretreatment time can be relatively short. Table 1: Matrices used for detection of toxic compounds. Matrix Collection procedure Samples pre-treatment Possible problems Blood Minimal Small sample volume Urine Invasive; requires veterinarian Non-invasive Extensive Metabolism Sweat Non-invasive Minimal Small sample volume Saliva Non-invasive Minimal Hair Non-invasive Extensive Small sample volume, external contamination External contamination Acute intoxication can take place on purpose or by accident (1, 2, 6-9). For example, in the Netherlands and some other countries in Europe intoxication of dogs is a well-known phenomenon (1). This can occur accidentally by eating e.g. rodenticides or can occur deliberately by poisoning (10). Also whole groups of animals can be intoxicated by the intake of feed which is contaminated with a toxic compound. This feed is contaminated by plant toxins or mycotoxins present in the field or on the plants during harvesting of the crops. An example of this is the intoxication of a flock of eight sheep by Pieris japonica in Belgium in 2005 (11). The sheep ate the leaves of the plant which were lying on the ground after trimming tasks in a neighboring garden. The sheep showed marks of poisoning such as salivation and five of the eight sheep died. Another recent example was the intoxication of a group of horses in The Netherlands (12). Multiple horses got sick and one horse died. After analysis of the horses organs and the feed at the farm, intoxication with colchicine was proven. Colchicine is a toxin which derives from the plant Colchicum autumnale (autumn crocus, meadow saffron or naked lady). The toxin came into the feed of the horses by mixing with the grass during harvesting. 1 Table 2 shows the number of reported cases of intoxication of animals from 2008 till 2013 in the Netherlands. The majority of them are related to pets and then specific dogs and cats. As can be seen in table 2, the total number of reported animal intoxications increased during the last years. This increase is probably due to an increasing report rate by animal owners (9). Table 2: Reported intoxications of animals in The Netherlands (9). 2008 2009 2010 2011 2012 2013 Dog 64% 63% 67% 68% 68% 70% Cat 23% 20% 23% 23% 24% 24% Horse 4% 2% 2% 1% 1% 1% Rabbit 2% 2% 2% 2% 2% 2% Cow 2% 7% 2% 3% - - Sheep 1% 1% 1% - - - Goat 1% 1% - - 1% - Bird 1% 3% 1% 1% 1% 1% Others 1% 1% 2% 2% 3% 2% Total # of animals 2917 3928 3424 3879 4205 4479 The examples mentioned above were all cases where it took some time until the source of the intoxication was discovered. The animals were already really sick or even died before appropriate action was undertaken. A reason for this can be the need of assistance by a veterinarian to take blood samples for investigation. In human toxicology the use of saliva as a matrix for analytical investigations is already accepted for many years (13, 14). Traditionally, saliva is mainly used for the analyses for illicit drugs such as cocaine and amphetamine (15). Also hormonal research has been performed using saliva; for instance steroid hormones (16). In the past decades a new type of analysis on the basis of saliva was introduced, namely DNA analysis, which is used for identification purposes (17). Saliva therefore is a widely used matrix in analytical human forensic toxicology. The aim of this research project is to develop a multi-residue method for the analysis of animal saliva using Liquid Chromatography – Mass Spectrometry (LC-MS). The development has to cover the entire procedure, from saliva sampling on location, e.g. at the farm to MS data interpretation. The method has to be able to deal with small amounts of sample material, because saliva sampling methods provide relatively low volumes of sample material. Acute intoxications occur at concentrations in the order of the median lethal dose (LD50). The LD50 is the dose of a compound where half the members of a tested population die. The LD50 for the majority of toxic compounds is in the mg/l range. This means the limits of Detection (LOD’s) of the compounds in the developed method don’t have to be in the μg/l (ppb) range, but rather in the mg/l (ppm) range. Compound classes of interest are plant toxins, mycotoxins, pesticides and veterinary drugs. Plant toxins and mycotoxins can enter the body of an animal by direct consumption of the plant or fungus or, more likely, by accidental mixing of toxic plants or fungi with other feed. Pesticides are the most likely substances used for deliberate intoxication by mixing a pesticide with the animals feed or water. Pesticide intoxication can also occur accidentally when animals eat for example rodenticides, which are not stored in a correct way. Veterinary drugs can be a cause of intoxication when an overdose has been 2 administered. The goal for this this research project was to develop a method capable to analyze more than 200 compounds of the classes mentioned before in samples of saliva in one LC-MS run. The method will be validated according an in-house validation protocol based on official international legislation. To our knowledge, no multi-class compound analytical toxicological screening method for veterinary saliva has been developed so far. As model animal saliva, saliva of calves will be used, because the availability of calf saliva for this research is significantly higher compared to that of other animals. 3 2 Background information 2.1 Saliva Saliva is an important element in the digestion process of a lot of animals. Some species even use saliva for nest building purposes (birds) and as a hunting tool where the saliva is venomous (vipers, cobras). In mammals saliva has various functions (13). It works as a lubricant for the mouth and gullet to make sure food passes easily. Furthermore, is helps moistening the food and creates a bolus which can be swallowed more easily. Saliva contains the enzymes amylase and salivary lipase which can break down respectively starch and fat in the food. Due to these enzymes the digestion already takes place in the mouth of the animal. Saliva also acts as a buffer for the mouth area to protect for example teeth for acidic conditions. Other functions where saliva plays a role are taste and microbial control. Figure 1 shows a scheme with the different functions of saliva in mammals. Figure 1: Functions of saliva (from Lamy et al. (18)). Saliva is produced in the salivary glands (13). Mammals have different glands in the mouth area where they produce saliva (see figure 2). Therefore one can distinguish between gland specific saliva and whole saliva. For certain purposes it can be useful to sample gland specific saliva when it is known that some substances of interest are produced in a certain gland. However, most saliva purposes require whole saliva. Whole saliva is a mixture of pure saliva from the salivary glands and other substances which occur in the mouth area. These can be bronchial and nasal secretions, blood from oral wounds, bacteria, viruses, fungi and food remains (19). The amount of saliva produced depends strongly on the animal. Human salivary production is on average between 1 and 1.5 liter per day, for horses it is up to 40 liter per day and for cows between 110 and 180 liter per day. Saliva production varies throughout the day and depends on whether or not an animal is resting or eating and the nature of the diet (18). The high rate of salivary production in cows can be explained by the fact that cows are ruminant animals. The pH of saliva in mammals varies between 5.5 and 8.5, dependent of the species and moment of salivary 4 production. For example, the pH of saliva from a cow is around 8.2, because of high sodium bicarbonate levels. The pH of saliva can be of high importance for the transportation of compounds between blood and saliva, as is explained in more detail in chapter 2.2. Figure 2: Schematic diagram of the salivary glands of four domestic mammals (from www.ucd.ie). 2.2 Transportation of toxic compounds in saliva There are two ways in which toxic compounds can enter the saliva of an animal. The first one is direct contamination of the saliva in the mouth area. This occurs when an animal is intoxicated orally, by eating intoxicated products or when intoxicated products are administered. The second way of contamination is by transport of compounds via the blood (5). Compounds enter the body orally or via intramuscular administration, for example injection of a veterinary drug. After oral administration, uptake of the compounds in the blood takes place in the digestion system. Primary organs where this uptake takes place are liver, stomach and the intestinal canal. Via the blood, compounds can be transported to the saliva. This transportation can occur in several ways and is dependent of the different chemical and physiological properties of the toxic compound and the saliva (20). Lipid solubility, degree of ionization of the compound in combination with the pH of the saliva and protein binding are the most important properties. Toxic compounds can enter the saliva via the lipid membrane by active or passive diffusion; active diffusion via pores in a membrane or passive diffusion via a concentration gradient. The presence of the 5 majority of toxic compounds results from passive diffusion (21). To represent the concentration ratio between saliva and plasma a so called S/P ratio is assigned. A ratio close to 1.0 means that the concentration of the compound in saliva and plasma are almost the same. A low value of the S/P ratio means the concentration in the plasma if higher than in the saliva, a high ratio means the concentration in the saliva is higher than in plasma. When a compound has a low S/P ratio, the chance of detecting it in saliva is not very high. Examples of toxic compounds with a relatively low S/P ratio are the pesticide atrazine (S/P = 0.66)(22), digoxin (S/P = 0.53) and diazinon (S/P = 0.16) (21). With the low S/P ratio of diazinon (0.16) there is still a significant concentration which can be found in saliva. 2.3 Toxic compounds The compounds of interest in this study are compounds which have a toxicological effect on animals and are available for intake by these animals. This intake can either be by the animal itself (eating, foraging) or by administration due to human interference. Component groups of interest are pesticides, natural toxins and veterinary drugs. Pesticides are used worldwide and on large scale to prevent vermin and diseases in for example crops. Intake of pesticides by animals can be accidentally or on purpose. Animal feed can be intoxicated with pesticides, animals may have access to pesticides which are not stored in a correct way or used for other purposes such as mouse or rat poisoning. Additionally, animals can be deliberately poisoned with pesticides in a conflict situating between persons (23). A well-known group of toxic compounds in the animal toxicology are the natural toxins. These are toxins which are produced by plants, fungi or animals. Some of these toxins are extremely toxic and are even used in the human crime world for poisoning purposes. Incidents with animals occur when they eat these toxic plants or when these plants or fungi are mixed in the animal feed. A third group of toxic compounds which are included in this research work are veterinary drugs. Veterinary drugs can be dangerous for animals when they are over dosed or when animals are extremely sensitive for the drug. Mostly intoxication by drugs is due to accidental intake. Table 3 shows the toxic compound classes which caused intoxications of animals in The Netherlands from 2008 till 2013 (9). Table 3: Toxic compound classes which caused intoxications of animals in The Netherlands (9). 2008 2009 2010 2011 2012 2013 Pesticides 24% 19% 21% 20% 21% 18% Human drugs 20% 19% 21% 22% 22% 23% Plant, fungi and animal toxins Household chemicals 19% 14% 18% 20% 20% 21% 12% 11% 10% 14% 12% 11% Food and drinks 7% 8% 10% 9% 10% 12% Industrial products 4% 10% 3% 2% 3% 3% Vet drugs 3% 5% 4% 5% 5% 5% Others 11% 14% 13% 8% 7% 7% 2.4 Liquid chromatography Chromatography is a technique which can separate different compounds in gas or liquid phase from each other. Gas phase chromatography is called gas chromatography (GC), where liquid phase chromatography is called liquid chromatography (LC). Different types of GC and LC have been developed, from simple techniques such as thin layer chromatography (TLC) to highly advanced 6 techniques such as ultra high performance liquid chromatography (UHPLC)(24). All chromatographic techniques work with the same basic principle of separating compounds due to differences in molecular characteristics in the mobile phase and the stationary phase in a chromatographic system. This stationary phase can be for example paper, C18 particles or a coated capillary. The separation characteristics can be for example molecular size, polarity or the ionization status of the molecule. In this research a variant of high pressure (performance) liquid chromatography (HPLC) has been used. HPLC is a separation techniques were compounds are separated on a column packed with stationary phase particles. The sample is injected in the flow of a liquid (mobile phase) which is pumped under high pressure through an analytical column. Due to the difference in affinity of the compound with the stationary and the mobile phase, the compound will stay on the column or will be transported through the column by the mobile phase. Because different compounds interact with the stationary and mobile phases differently, a compound can be separated from matrix compounds or from other compounds of interest. To achieve optimal separation different stationary phases and mobile phases can be used. The mobile phase can be used in an isocratic or gradient flow. When using isocratic flow, one eluent is used by using a single pump. This eluent can consist of multiple fluids in a fixed composition, e.g. 60% water and 40% methanol. With a gradient flow a binary pump is used, so different eluents can be mixed to create a variable eluent composition in time. Advantage of a gradient flow is that analysis time is reduced and samples which cannot be separated with an isocratic flow can be separated better due to the increase in eluent strength by changing the eluent composition. For example a gradient flow can start with an eluent composition of 90% water and 10% methanol and can change to 100% B in for example 10 minutes time. To achieve better separation (higher resolving power and peak capacity) smaller stationary phase particle are needed (25). This can be explained using the Van Deemter curve, which shows the correlation between the resolving power (Height Equivalent Theoretical Plate, HETP) of the column and the mobile phase flow rate (26). The better the resolving power, the lower the value for HETP and at the minimal HETP the flow rate is optimal. In UHPLC, using smaller particle sizes compared to HPLC, the part of the curve on the right of the minimum is much lower (see figure 3). This means that higher mobile phase flow rates can be used while retaining optimal resolving power. To achieve these higher flow rates the LC systems need to be able to provide high pressures. Figure 1: Van Deemter curves describing the dependence of the plate height (H) on the linear velocity of the mobile phase (u) for stationary phases with different particle sizes (26). 7 Because sample volume, in combination with sensitivity, is important in this research project a microUPCL has been used. With microUPLC the inner volumes of the LC tubing are smaller than with conventional UPLC, so, to achieve the same amount of sensitivity, less sample volume is needed or with the same sample volume a significant gain in sensitivity can be achieved. The gain in sensitivity resulting from the use of a microUPLC column compared with a conventional UPLC column is shown by the following relation: Formula 1 where f is the gain in sensitivity, d1 and d2 are the diameters of the conventional UPLC and microUPLC columns, respectively (27). This means in theory that reducing the column internal diameter form 2.1 mm to 150 μm, should result in an almost 200-fold gain in sensitivity. Also the flow rate is much lower (1-5 μl/min), so a significant reduction of mobile phase can be achieved, which reduces costs and is ecologically sustainable. A disadvantage of nano- and microUHPLC systems is the complexity of use. Combining tubing or capillaries and nano- or micro columns to each other in the right way can be very time consuming. Furthermore, because the flow rate is very low it is difficult to detect possible leakages. Therefore, manufacturers try to come up with inventive techniques to avoid these problems. One of these inventions is the chip based LC column system. In these systems the analytical column is build inside a chip, so no manual connections have to be made. The system which has been used for this research project uses the IonKey technology from Waters Corporation. The research was performed with a test-version of the system, because RIKILT and Waters Corporation have an agreement for testing this new technology (28). By integrating a microLC column in a chip which can be slided into the source of the mass spectrometer, leakages by connections are minimized. IonKey is a chip based microUPLC technique, which uses a so called tile or iKey which has to be placed into a special designed electro spray ionization source (see figure 4). 8 Figure 2: IonKey technology with the iKey inside the IonKey source. The iKey consists of an inlet (from microLC pump or syringe pump) connected to the packed column or a capillary. An iKey with a capillary can be used for infusion experiments, for example the calibration of the mass spectrometer. The column can be packed with different stationary phase materials, all C18 based. BEH (Ethylene Bridged Hybrid), CSH (Charged Surface Hybrid) and HSS (High Strength Silica), stationary phases are available that can be used for different types of compounds depending on the application. At the end of the column there is an electro spray ionization emitter. Because of the design of the iKey only positive ionization mode is possible yet. Figure 5 shows a schematic overview of the iKey. The electrospray ionization emitter works similar as a traditional electrospray ionization source. Figure 3: iKey schematic view with the different inlets, the actual analytical chromatographic column and the electrospray emitter (from www.waters.com). When optimizing the working of the IonKey some parameters can be varied. These are the position of the iKey, the capillary voltage, the capillary gas flow and the capillary temperature. The position of the iKey can be altered in the x, y and z direction. Because of the low eluent flow it is important to accurately 9 set the position of the iKey. For this purpose a camera is installed on the source. With this camera images of the sample flow and lock mass flow can be viewed, so the position of the iKey towards the entrance of the mass spectrometer can be optimized. The capillary voltage, capillary gas flow and temperature are important for an optimal evaporation and ionization of the sample flow. The settings for these parameters are dependent on the sample flow. Generally the higher the flow, the higher the values have to be. Appendix I shows a picture of a camera shot at the inside of the IonKey source. 2.5 Mass spectrometry For screening purposes Time of Flight (TOF) mass spectrometry is an often used technique. A screening method is a method where samples after analysis can be categorized as compliant (negative) or potentially non-compliant (suspected). When a sample is compliant no compounds of interest are found and the sample can be regarded as negative. When the sample is non-compliant one or more compounds are detected in the sample. For screening methods this means the sample is suspected and an additional confirmation analysis has to be performed to confirm the identity of the compound. Time of Flight mass spectrometry is used to measure compound masses with a higher accuracy compared with for example triple quadrupole mass spectrometers. Not the mass of an ion is measured, but the ratio between the mass and charge of the ion (m/z). When the charge of the ion is singular, the exact mass is measured. When for example the charge of the ion is double, half the exact mass of the ion plus two protons is measured. As the name already implicates, TOF uses the time an ion needs to travel a certain distance through a flight tube to separate ions with different masses form each other (29). This TOF tube can consist of one single tube or one or more so called reflectors. These reflectors ‘reflect’ the ions back into the tube and in this way doubles or quadruples the path length and also corrects for thermal energy differences in ions with the same mass to charge ratio (see figure 6). Due to the longer flight path the accuracy and resolution will improve. For detection in a TOF instrument mainly a microchannel plate detector or a secondary emission multiplier (SEM) is used. With TOF only full scan spectra can be acquired, so no precursor selection and no fragmentation data can be obtained. When using a QTOF system precursor selection and fragmentation is possible. A QTOF is a hybrid system where a quadrupole is placed before the TOF tube (see figure 6). This quadrupole can be used for pre-cursor selection, so specific ions can be selected and transferred to the TOF tube. Behind the quadrupole a fragmentation cell is placed, so the selected pre-cursors can be fragmented and these fragments can be transferred to the TOF tube. In this research measurements were done with a Waters Xevo QTOF. This instrument has a method option called MSe, where alternating full scan and fragmentation scan spectra are acquired. These spectra can be linked to each other based on scan time, so for every time point in the chromatogram there will be a full scan spectrum and a fragmentation scan spectrum. This fragmentation scan is an all ion fragmentation scan, so all ions which are passing the collision cell are fragmented. This fragmentation is performed using a stepped collision energy, which means that during every fragmentation scan different collision energies are applied on the ions. In this way ions which fragment with lower energy as well as ions which fragment with higher energy will be fragmented. The instrument has a Lock Mass correction device. This is an external syringe pump which introduces a lock mass solution via a separate capillary into the ESI source. Via a baffle, a plate which switches between sample and lock spray, every twenty seconds (or another interval which can be chosen by the operator) the lock spray will be introduced. This lock spray solution consists of a compound with known mass or masses. These masses are detected and the deviation between the exact mass and the 10 measured mass is calculated. Directly during the analysis or during data processing all the other measured masses are corrected with this calculated deviation. In this way the instrument corrects for small deviations in measured mass which is caused by for example temperature changes in the environment. The capillary and baffle of the lock spray device can be seen in Appendix I. Figure 4: Schematic view of a QTOF MS with from left to right the ionization source, quadrupole, collision cell and a W mode reflectron TOF tube (from giga.ulg.ac.be). The TOF MS has to be calibrated on a regular base, depending the standard practice of the company or user. This calibration has to be performed to make sure the instrument is capable of performing according to the specifications of the instrument or the company. A calibration solution is introduced into the MS by infusion with an internal or external pump. This solution contains compounds with known exact masses, which are present in a calibration table. The measured exact masses are compared with the known exact masses and the deviation is calculated. This deviation has to fall within a pre-defined criterion, for example < 2 mDa. When for all tested masses this criterion has been achieved the calibration can be accepted and the TOF MS is calibrated and can be used for analysis. 2.6 Data handling Mass spectrometry data is acquired using Waters Masslynx software. Further data processing was performed using UNIFI. UNIFI is a new software package of Waters which is still in the development phase. Masslynx has already an option to correct the measured mass with the help of the lock mass when analyzing. When selecting this option the data has already been corrected before processing with UNIFI. In UNIFI a targeted library has been build including exact masses and retention times of all compounds included in the standard mixture. If known also common fragment masses of the compounds are included in the method. The retention times were determined by injection of standard solutions containing all compounds. When multiple peaks were present in the chromatogram, the MSe function of the acquisition method was used to identify the correct peak using the product ions of the compound. The retention time window used in the processing methods was 0.1 minute, which means the software only looks at peaks 3 seconds before and 3 seconds after the entered retention time. Mass accuracy was 11 set at 25 mDa. This means the software only looks at masses which are in between 25 mDa of the entered exact mass. Different adducts can be included in the method, for this study H+ and NH4+ adducts were included. 2.7 Validation To test the performance of the developed method, the method has to be validated. This means that different performance characteristics have to be determined. Validation of analytical methods has to be performed according to national and international legislation. For different research fields, different official validation documents were published. Since this research was performed within the RIKILT Business unit Veterinary Drug Research, the validation was performed using EU Commission Decision2002/657/CE (30). This document describes which characteristics should be determined for the validation of an analytical method in the veterinary drug residue research field. The document distinguishes between confirmatory and screening methods and between qualitative and quantitative methods. Figure 7 shows the performance characteristics which should be determined when validating one of the methods mentioned above. Figure 5: Classification of analytical methods by the performance characteristics that have to be determined according Commission Decision2002/657/CE (30). Before starting the validation it has to be determined in which way the method has to be validated. The method which has to be validated in this research project is a qualitative screening method. So according to the table the following characteristics have to be determined: Detection limit (CCß), selectivity/specificity and applicability/ruggedness/stability. These characteristics will be explained below. Detection limit (CCß): The detection limit is the limit where with a certain certainty (ß) can be stated that the found value is non-compliant (30). In this validation ß correspond with a certainty of 95%. This means that a found concentration above this CCß limit can be stated as a suspect sample (31). Selectivity/specificity: Selectivity or specificity says something about the statement how certain one is that the compound found really is that compound. So the more certain one is that the compound found is 12 really that compound the more specific or selective the method is. LC-MS already is a selective technique, because it measures retention times in combination with exact masses, which are specific for a certain compound. To verify the selectivity blank saliva samples with and without addition of the compounds will be analyzed. When no compounds are identified at the specific RT and mass of these compounds in the saliva samples the method is selective for these compounds. Ruggedness: The ruggedness (robustness) of the method says something about the capability of the method to deal with certain changes in the method. To test this certain steps in the method which could be critical are tested. When the results (number of identified peaks) of the ruggedness samples are comparable with those of the control samples the method is robust for this step in the method. Stability: The stability refers to the stability of the measured compounds in standard solutions and/or final extracts. The stability of standard solutions is tested by comparing standards which have been stored for a certain time in the refrigerator or freezer with standard which have been stored at -80°C. The -80°C samples are assumed to be stable. After a certain storage time the standards are analyzed and the results are compared. When the results are comparable (e.g. < 10% deviation in the measured peak area) the compound is considered to be stable for the storage time and condition. Stability of extracts is determined by storing sample extracts for a certain time (e.g. one week) in the refrigerator or freezer before re-analyzing and compare the results with the original analysis. When the results are comparable the extract are considered to be stable for the storage time and condition. The way the above mentioned validation characteristics were determined is described in an in-house validation plan. This plan is approved by the quality assurance officer before the validation can take place. 13 3 Materials and Methods In this chapter the materials used, chemicals and apparatus will be explained. The methods which have been used for sample collection and sample pre-treatment are explained. Also the animals which have been used for the collection of the saliva samples will be mentioned in this chapter. 3.1 Materials Animals: The saliva samples were collected from calves. For the preliminary sampling tests calves were used from the dairy farm of Mts Meijer-Pelgrum in Lettele, The Netherlands, were samples were taken from calves in the age of 4 weeks till 3 months. For the validation study samples were taken from the same farm from calves in the ages of 2 till 3 months. These calves were fed milk, hay and maize during the day and were able to drink water whole day. For the sampling tests and the validation study calves were samples at Zodiac Carus. This is an animal research facility of Wageningen University and Research centre. The animals were 2-4 months old and were fed hay and maize during the day and able to drink water whole day. All animals were kept in spacious sheds, according the Dutch law on animal housing (activiteitenbesluit en activiteitenregeling, Dutch Ministry of Infrastructure and Environment). Chemicals: Acetonitrile, methanol, formic acid and water were al ULC grade and were purchased from Actu-All Chemicals (Oss, The Netherlands). DMSO, Sodium hydroxide and 2-propanol were purchased from Sigma-Aldrich (Saint Louis, MO, USA). Reagents: Mobile phase A (Water/Acetonitrile/Formic acid (99 / 1 / 0.1) (V/V/V)), Mobile phase B (Acetonitrile/Water/Formic acid (99 / 1 / 0.1) (V/V/V)), Water / 2-propanol ((1 / 1) V/V)), Leucine/Enkypheline (2000 μg/l), MS Leucine/Enkypheline Kit (Waters, 700002456) Standard solution: All standard solutions were made according in-house standard operating procedures. Standard mixtures of the different compound groups were made in concentrations of 50 mg/liter. Table 5, 6 and 7 show the compounds present in the standard mixtures. The three mixtures are combined in two Tox mixtures, Total Tox mix 10 mg/l and Total Tox mix 1 mg/l. These two mixtures are being used for the additions in the validation study. Furthermore there is an internal standard mixture where clenbuterol-d6 and C- 13 caffeine (Caffeine-(3-methyl-13C)) are present in a concentration of 10 mg/l Veterinary drug mixture 50 mg/l Natural toxin mixture 50 mg/l Pesticide mixture 50 mg/l Total Tox mix 10 mg/l 14 Total Tox mix 1 mg/l Internal standard mix 10 mg/l Table 5: Toxins. Component Component Component Component Abrine Digitoxigenin Lupanine Rottlerin Aconitine Digitoxin Lupinine Santonin Aflatoxin B1 Digoxin Lycopsamine Scopolamine Aflatoxin B2 Echimidine Monocrotaline Senecionine Aflatoxin G1 Emetine Morphine Senecionine-N-oxide Aflatoxin G2 Ephedrine Narceine Seneciphylline Ajmalicine Erucifoline Noscapine Seneciphylline-N-oxide Anisatine Erucifoline-N-oxide Ochratoxin A Senkirkine Aristolochia acid I Genistein Solanine alpha Aristolochia acid II Atropine Geranyloxypsoralen 5(Bergamottin) Grayanotoxin III Ouabain (Strophanthin G-) Papaverine Podophyllotoxin Strychnine 13C-Caffeine Heliotrine Pulegone Teucrin-A Colchicine Imperatorin Rescinnamine Convallatoxin Jacobine Reserpine Tetrahydrocannabinol (THC) Tropine Coumarin Jacobine-N-oxide Retorsine Tubocurarine Cucurbitacin I Kavain Retorsine-N-oxide Umbelliferone Cytisine Lobeline Ricinine Vincamine Component Component Component Component 13C-Caffeine diflubenzuron isoproturon propyzamide 2,4-D diflufenican isopyrazam prosulfocarb Abamectin dimethenamid Isoxaben pymetrozine Acephate Dimethoate isoxaflutole pyraclostrobin Acequinocyl dimethomorph kresoxim-methyl pyridaben Acetamiprid dinoterb Lenacil pyridate Aclonifen Diuron linuron pyrimethanil Aldicarb DNOC lufenuron pyriproxyfen ametoctradin dodemorph Malathion pyroxsulfam Amisulbrom dodine mandipropamid quinmerac Asulam emamectin MCPA quinoclamine Azadirachtin epoxiconazole MCPP Quinoxyfen azamethiphos Ethirimol mepanipyrim quizalofop-ethyl azoxystrobin ethoprophos mesosulfuron-methyl rimsulfuron Bendiocarb etoxazole mesotrione silthiofam Bentazone famoxadone metalaxyl Simazine Bifenazate fenamidone metamitron spinosyn-A Bifenthrin Fenamiphos metazachlor spinosyn-D Bixafen fenhexamid metconazole spirodiclofen Boscalid fenoxaprop-ethyl Methabenzthiazuron spiromesifen brodifacoum fenoxycarb Methamidophos spirotetramat Sparteine Table 6: Pesticides. 15 bromadiolone fenpropidin methiocarb Spiroxamine Bromoxynil Fenpropimorph Methomyl sulcotrione Bupirimate fipronil methoxyfenozide tebuconazole Buprofezin flonicamid metolachlor tebufenpyrad Carbaryl florasulam Metoxuron teflubenzuron carbendazim Fluazifop metrafenone tembotrione carbetamide fluazinam metribuzin tepraloxydim Carbofuran Flucycloxuron metsulfuron-methyl terbuthylazine carfentrazone-ethyl fludioxonil Mevinphos Terbutryn Chlorantraniliprole Flufenacet Myclobutanil Tetraconazole Chlorbromuron flufenoxuron nicosulfuron thiabendazole Chloridazon fluopicolide Omethoate thiacloprid clodinafop-propargyl fluoxastrobin oxamyl thiamethoxam clofentezine fluroxypyr Oxydemeton-methyl thiophanate-methyl Clomazone flutolanil paclobutrazol Tolylfluanid Clopyralid foramsulfuron penconazole topramezone Clothianidin fosthiazate pencycuron tri-allate Cyazofamid Haloxyfop Phenmedipham tribenuron-methyl Cybutryne Haloxyfop-p-methyl picoxystrobin triclopyr Cymoxanil hexythiazox pinoxaden trifloxystrobin cyproconazole imazalil pirimicarb triflumizole Cyprodinil imidacloprid pirimiphos-methyl Triflumuron Cythioate indoxacarb prochloraz triflusulfuron-methyl desmedipham iodosulfuron-methyl Profenofos Triforine dichlofluanid ioxynil propamocarb trinexapac-ethyl difenoconazole Iprovalicarb propiconazole tritosulfuron Table 7: Veterinary drugs. Component Component Component Component Chlortetracycline Abamectine Clencyclohexerol Salmeterol Demecloxycline Doramectine Clenpenterol Terbutaline Doxycycline Emanmectine Clenproperol Tulobuterol Oxytetracycline Eprinomectine Fenoterol Zilpaterol Tetracycline Ivermectine Isoxsuprine Clenhexyl Lasalocid Moxidectine Mabuterol Ritodrine Maduramycine Broombuterol Mapenterol Metaproterenol Monensin Carbuterol Procaterol Hydroxymethylclenbuterol Narasin Cimaterol Ractopamine Bromchlorbuterol Salinomycine Cimbuterol Reproterol Semduramycine Clenbuterol Salbutamol Apparatus: Standard laboratory equipment was used for sample preparation and analysis. These consist of: Saliva sampling devices (animal cotton rope, Salimetrics)(Swab, Henry Schein), Centrifugal tubes for saliva sampling device (custom made), Centrifugal tubes for saliva sampling device (Salimetrics), Tubes 50 ml (Greiner), Nitrile gloves (Medica), Tubes 1.5 ml (Eppendorf), Tube 12 ml with screw cap (Greiner), Vial 16 with screw cap, total recovery (Grace), Glass flasks, 250 ml (Scott), Pipettes of different types and volumes, each calibrated according RIKILT SOP, Centrifuge (Eppendorf), Vortex (IKA). LC-MS system consisting of: NanoAcquity UPLC (Waters Corporation), IonKey MS source (Waters Corporation), iKey BEH C18 Separation Device, 130Å, 1.7μm, 150 μm x 50mm (Waters Corporation), Xevo QTOF Mass spectrometer (Waters Corporation). Settings of the different systems can be found in table 8 and 9. Table 8: LC and IonKey settings. Flow 3 μl/min Gradient See table 12 Injection volume 2 μl Capillary Voltage 3.5 kV Nanoflow gas 0.5 bar iKey temperature 45°C Sample tray temperature 8°C Table 9: QTOF MS settings. Source type Electro spray ionization – positive mode Mass range 50 – 1250 Da Scan time 0.2 seconds Cone voltage 20 kV Source temperature 120°C Cone gas flow 30 l/hour MSe collision energy ramp 15 – 40 V 3.2 Methods 3.2.1 Sampling devices Saliva sampling devices for human collection are commercially available on a large scale. Examples of these are cotton buds (swabs) and spitting tubes of different companies. Devices for animal use on the other hand are not so common. The reason for this is the fact that animal saliva is not used a lot as a matrix of investigation. However, a few saliva sampling tools especially designed for animals are commercially available. There is the animal cotton rope by Salimetrics and the pig cotton rope (Happy Bite) designed by GD animal health in the Netherlands. Because these were the only animal sampling devices found also human sampling devices were tested and used in this research. For the testing some factors were important: ease of sampling, absorption capacity of the device and ease of saliva extraction from the device. Most (human) saliva devices come with a manual where a short the sampling procedure is explained. Important is to avoid contamination of the device, so gloves must be worn during sampling (different for every animal) and the devices must be put directly after sampling in a storage tube. This can be part of the sampling device (for some swabs) or this can be a separate tube. Another important factor is the sampling time; the time the device stays in the mouth of the animal. For most devices it was stated that the device should stay in the mouth for a time period of one minute. 17 Figure 8: Pictures of two sampling devices: swab (left) and cotton rope (right). 3.2.2 Salivary collection Saliva collection has to be performed by one person when the calf can be fixed in the feeding fence or by two persons when the calf cannot be fixed in a feeding fence. One person fixes the calf in a feeding fence while the second person performs the actual sampling (figure 9). The mouth of the calf has to be held open and the sampling device has to be placed in the mouth. The device has to be kept in the mouth for a certain time before it can be taken out again. The device is transferred into a coded tube and stored in a cooled environment until it is taken to the laboratory. Figure 9: sampling of saliva from a calf. One person fixes the calf, the other performs the actual sampling. 18 3.2.3 Sample pre-treatment After sampling the samples have to be stored in a freezer (32) or an extraction step (extraction of the saliva from the sampling device) has to be performed. The extraction of the saliva from the device is done by centrifugation. Depending on the used device the centrifugation step can differ slightly. Using a rope kind of device, the device has to be cut in pieces of 3-5 centimeters and put in a 50 ml centrifugal tube with an centrifugal insert (filter). Using swab kind of devices, the actual swab has to be cut of the stick and transferred to a 10 ml centrifugal tube with an centrifugal insert (filter). The tubes are centrifuged for 10 minutes at 3200 g. The saliva which is at the bottom of the tubes is transferred to a clean tube. Before the saliva can be analyzed there has to be a sample pre-treatment procedure. The main reason for this is to remove the proteins (precipitation) and feed and dirt particles which are present in the saliva. Some different methods were testes: 1. No protein precipitation. 2. Protein precipitation using acetonitrile and methanol 3. Filtration of the saliva to get rid of the proteins 4. Combinations of 2 and 3 Results of the different pre-treatment methods can be found in chapter 4.2. 3.3 Validation The validation has to be performed according an in-house validation protocol. This protocol is based on EU legislation (see paragraph 2.7). The validation is carried out during three days. Every day three times seven different blank saliva samples are used for the experiments. One set of seven blank samples is only fortified with internal standard mixture. These samples are used for the determination of the selectivity. The other two sets of seven blank samples are fortified with standard mixtures of the toxic compounds at two different concentrations. These concentrations are 100 microgram per liter and 1000 microgram per liter. These samples are used for the identification of the compounds and the determination of CCß. At one of the three days 6 additional samples (of one blank saliva sample) are analyzed for the determination of the ruggedness. A scheme of the experiments of validation day 3 is shown in appendix V. In this study the internal standards are add to the samples to verify if the sample pre-treatment went well. So if the internal standards are not identified in the sample after data processing, there was something wrong with the sample-pre-treatment in that sample. 19 Table 10 shows the scheme of the ruggedness test. Two steps are tested, shaking the sample during protein precipitation and addition of DMSO. When the results (number of identified compounds) of the ruggedness samples are the same as the control sample the method is robust for that step in the method. Table 10: Ruggedness of the method. Two varieties on the method were tested, shaking during protein precipitation and DMSO addition. SOP (A) Variable (B) Day 3 Level (duplo) 0.5 mg/l Criteria According to SOP According to SOP Day 3 0.5 mg/l Shake 30 sec vortex Shake 10 sec vortex 95% Peak identification Day 3 0.5 mg/l 5 microliter DMSO No DMSO 95% Peak identification The stability of the extracts is determined by storing the extracts of one of the validation days in the freezer for one week. After this week they are re-analyzed using the same analyzing method. When the results of the stability samples are comparable with the results of the original analysis, the samples are considered to be stable for one week in the freezer. 20 4 Results and discussion In this chapter the results will be presented and discussed. Sampling, sample pre-treatment, LC-MS analysis and validation will be discussed in four different paragraphs. 4.1 Sampling Different saliva sampling devices were tested within this project. A distinction can be made between small volume and large volume devices. The small volume devices can be used for collecting volumes less than 300 µl, the large volume devices can be used for volumes larger than 300 µl. The small volume devices are the so called swabs, a stick with a cotton part on top. Different devices of different manufacturers were tested and assessed for amount of saliva uptake, user friendliness and costs. Taken these factors in account, the cotton swab of Henry Schein, (900-3156) is the most suitable small volume device. This device consists of a wooden stick with cotton wool top. The saliva can be sampled and the top of the stick can be broken of the rest of the stick and placed into a centrifugal device. The samples should be prepared according paragraph 3.2.3 or placed in the freezer immediately. Three large volume sampling devices were tested, all cotton ropes of different sizes. Table 11 shows the different sample volumes which were collected with the different ropes. Table 11: Saliva yield of the three different tested large volumes sampling devices. Device Sample # Saliva (± µl) Cotton rope Custom rope Children swab 1 2 3 1 2 1 2 3 3500 400 700 700 2000 1700 1000 3500 The so called ‘children swab’ showed good saliva yield, but is too small to put in an animals mouth without the risk of swallowing. The custom made rope also showed good results concerning saliva yield. The problem with this device is that it takes some time to manufacture it and the that cotton wool which is used comes of the device, so the animal can swallow it. The most suitable large volume device, taken into account the 3 factors mentioned above, is the animal cotton rope of Salimetrics. The saliva yield is good (most samples > 250 μl), it can be held firmly by the sampler and the costs are relatively low. Table 12 shows the saliva yield of the animal cotton rope sampled at three different days for nine different calves. Table 12: Saliva yield of the cotton rope device at three different sampling days (3,4 and 5) for nine different calves (2-18, even numbers). sample 3.2 3.4 3.6 3.8 3.10 3.12 3.14 3.16 3.18 saliva (± µl) ±650 ±115 ±1000 ±450 ±330 ±330 ±900 ±1700 ±620 sample 4.2 4.4 4.6 4.8 4.10 4.12 4.14 4.16 4.18 saliva (± µl) ±2350 ±1200 ±100 ±250 ±2250 ±1050 ±750 ±700 ±1950 sample 5.2 5.4 5.6 5.8 5.10 5.12 5.14 5.16 5.18 saliva (± µl) ±1250 ±600 ±500 ±600 ±1000 ±250 ±330 ±800 ±950 As can be seen in table 11 the variation of saliva yield of the different samples is large. This is due to the different calves which were sampled. The saliva production during the day varies a lot, especially with ruminating animals. 21 Figure 10: Saliva sampling from a calf with one of the tested swabs. 4.2 Sample pre-treatment Different sample pre-treatment procedures for saliva clean up were tested: 1. No protein precipitation, only dilution 2. Protein precipitation using acetonitrile and methanol 3. Filtration of the saliva to remove proteins 4. Combinations of 2 and 3 The procedures were tested with saliva samples from calves (see paragraph 3.1). The samples were fortified with a test mixture of 43 pre-selected compounds of the three compound classes. Recoveries (identification and peak intensities) of the compounds were used to evaluate the procedures. For the first option, the samples were injected directly on the LC-MS system after a 10-times dilution with water. The LC-MS system gave an over-pressure error. The pressure on the IonKey exceeded the maximum (10.000 psi). This was probably due to clogging of protein on the column with the increasing percentage of organic solvent (acetonitrile) in the gradient. The column could not be used anymore, so no further experiments without protein precipitation were performed. For the second option, 100 microliter saliva was pipetted in an Eppendorf tube. 250 microliter of methanol or acetonitrile was added and the tubes were shaken on a vortex for 30 seconds. Then the tubes were centrifuged for 10 minutes at 14.000 g. After centrifugation, the supernatant was transferred to a total recovery LC vial containing 5 microliter DMSO. The sample was evaporated to almost dry (DMSO) at 40°C under a stream of nitrogen. 100 microliter water was added to the vial and shaken on a vortex for 10 seconds. During analysis, no clogging of the IonKey occurred, so it seems there was sufficient protein precipitation from the sample to prevent clogging. The results for acetonitrile and 22 methanol were similar, both solvents worked well. On the other hand, acetonitrile gave better chromatographic results compared to methanol. The background noise in the chromatograms was reduced using acetonitrile, so identification of the compounds was more straightforward. Figure 11 shows two extracted ion chromatograms (XIC) of clenbuterol of the same saliva sample: the chromatogram at the top after acetonitrile protein precipitation and the chromatogram at the bottom after methanol protein precipitation. It can be seen that both the intensity of the clenbuterol peak and the signal to noise ratio (S/N) is higher in the acetonitrile protein precipitation sample. This was the case with all of the tested compounds in the test mixture. X1_140513_SALTO_021 1: TOF MS ES+ 277.087 0.0500Da 286 5.32 100 % acetonitrile 5.88 10.15 4.64 0 1.00 2.00 X1_140513_SALTO_006 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 18.00 1: TOF MS ES+ 277.087 0.0500Da 93 17.00 18.00 1: TOF MS ES+ TIC 8.57e3 5.33 100 methanol % 5.29 8.68 5.86 6.22 4.674.85 1.27 1.67 0 1.00 2.00 X1_140513_SALTO_021 3.00 4.00 5.00 6.00 8.47 7.52 7.68 7.00 8.00 9.00 5.68 10.00 11.00 12.00 13.00 14.00 15.00 16.00 9.19 5.31 100 10.16 10.13 10.18 9.33 10.08 5.86 1.57 Figure 11: XIC chromatograms of clenbuterol with acetonitrile (top) and methanol (bottom) protein 15.31 precipitation. Using acetonitrile gives higher peak intensities 15.04 and lower S/N ratios compared to 15.41 8.66 8.79 7.72 8.33 methanol. 15.66 6.01 4.28 7.00 7.95 % 4.75 1.36 0.03 1.78 2.62 3.35 15.99 4.02 9.41 9.79 17.32 10.64 0.69 17.99 18.65 18.91 14.89 11.43 0 1.00 2.00 X1_140513_SALTO_006 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 18.00 1: TOF MS ES+ TIC 100 For the third option ultrafiltration was used. Ultrafiltration is a filtration technique9.53e3 where a sample is 1.58 1.36 1.28 filtered using centrifugal forces using special manufactured centrifugal filters. The idea is that large 6.20 7.99 1.13 % molecules (proteins) and possible dirt particles stay on the filter. The filtrate then is processed further. 4.77 1.08 0.49 2.02 2.67 2.82 4.30 5.32 6.01 6.34 15.04 7.45 7.75 8.15 8.44 15.45 9.35 15.74 Different types of ultrafilters were tested containing different filtermembranes, all with a 3.000 Da 3.79 16.43 10.63 10.95 11.44 17.32 17.52 18.76 18.91 14.89 0 Time membrane. VWR Centrifugal Filter PES 3K, Millipore Amicon Centricon YM 17.00 3K and Millipore Amicon Ultra 1.00 2.00 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 18.00 3K. The results looked promising, which means no clogging of the IonKey occurred and good recoveries (number of detected compounds) of the compounds that were added to the saliva were obtained. However, contrary to these good results in the first attempts, succeeding experiments still gave clogging of the IonKey, which most likely means the proteins were not removed from the saliva samples sufficiently. The most likely explanation for this observation is a variable protein concentration in the samples. Using only ultrafiltration therefore is not a good option. The fourth option tested was a combination of the techniques mentioned above. Combinations of protein precipitation and filtration were used as well as combinations of dilution and filtration. These options gave good results concerning protein precipitation, but there was no advantage compared to protein precipitation with methanol or acetonitrile alone. Reduction of background noise did not improve and there was no increase in identified compounds compared with protein precipitation with acetonitrile alone. Figure 12 shows an example of clenbuterol in saliva with and without ultrafiltration. The saliva sample without ultrafiltration (only precipitation with acetonitrile) has significantly higher intensity and a 23 higher signal to noise ratio. From this it can be concluded that using ultrafiltration causes loss in intensity or even loss off compounds due to interaction with the filter membrane. X1_140513_SALTO_023 1: TOF MS ES+ 277.087 0.0500Da 599 5.37 100 % acetonitrile without ultrafiltration 2.47 2.59 0.56 0.97 1.28 2.70 1.69 4.74 5.01 5.90 6.666.72 7.19 8.66 9.20 15.08 15.47 10.06 0 1.00 2.00 X1_140513_SALTO_008 3.00 4.00 5.00 3.09 3.08 3.11 100 6.00 7.00 8.00 9.00 10.00 3.05 3.13 9.749.80 % 2.97 9.68 9.64 9.58 5.24 3.20 5.44 2.92 1.71 2.82 13.00 14.00 15.00 16.00 17.00 18.00 1: TOF MS ES+ 277.087 0.0500Da 205 acetonitrile with ultrafiltration 3.00 3.16 0.51 12.00 5.36 3.03 0.50 0.66 1.26 1.65 11.00 4.70 3.38 8.66 5.57 6.18 6.69 4.78 9.53 8.97 9.92 9.98 10.02 15.04 10.12 10.36 10.45 15.12 0 1.00 2.00 X1_140513_SALTO_023 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 10.06 100 18.00 1: TOF MS ES+ TIC 3.31e5 Figure 12: XIC chromatograms of clenbuterol with and without ultrafiltration in the sample pre6.13 9.35 5.86 treatment. Using ultrafiltration gives lower10.33peak intensities and higher S/N ratios compared not 5.71 10.72 11.07 5.57 using ultrafiltration. % 6.55 6.64 6.75 1.38 1.59 0.62 5.02 5.22 1.66 4.75 0 1.00 2.00 X1_140513_SALTO_008 3.00 4.00 15.12 15.47 11.51 12.01 4.35 4.47 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 18.00 1: TOF MS ES+ TIC 100 2.45e5 Summarizing all the above it can be concluded that the optimal sample pre-treatment method was 6.02 6.57 6.66 6.30 7.20 protein precipitation using acetonitrile. The full sample pre-treatment procedure is described in Appendix 5.74 5.59 % IX. 10.27 10.46 5.03 5.24 1.51 1.61 10.63 0.60 4.79 8.18 8.47 4.41 9.53 11.19 14.89 3.11 0 Time 1.00 4.3 2.00 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 18.00 LC-MS analysis LC: Different eluent compositions were tested to achieve optimal chromatographic conditions. Because a large number of different compounds will be introduced, it is necessary to have a good chromatographic separation between all compounds. This will, however, mean that compromises have to made and some peaks won’t be as sharp as for a dedicated single compound LC method. Eluents using water, methanol an acetonitrile were tested. As additive, formic acid at a concentration of 0.1 % (v/v) was used. Figure 13 shows chromatograms of some veterinary drugs analyzed with an eluent consisting acetonitrile or methanol. The peaks in the extracted ion chromatograms (XIC) with acetonitrile are sharper and the intensities are higher compared to the use of methanol in the eluent. There is also significantly less noise in the acetonitrile chromatograms. The recoveries of the compounds (identified peaks) were similar when using acetonitrile or methanol. Based on these results an eluent composition consisting of water, acetonitrile and formic acid was chosen for the final method. 24 X1_140422_SALTO_006 10.44 100 1: TOF MS ES+ 416.3 0.0500Da 52 methanol XIC salmeterol % 10.40 10.38 10.92 12.54 12.86 13.09 14.09 14.40 14.66 0 1.00 X1_140422_SALTO_003 2.00 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 7.99 15.00 16.00 17.00 1: TOF MS ES+ 416.3 0.0500Da 61 16.00 17.00 1: TOF MS ES+ 277.09 0.0500Da 301 acetonitrile XIC salmeterol % 100 14.00 10.17 7.32 0 1.00 X1_140422_SALTO_006 2.00 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.52 10.00 11.00 12.00 13.00 6.70 100 14.00 15.00 methanol XIC clenbuterol % 6.68 6.38 0 1.00 X1_140422_SALTO_003 2.00 3.00 4.00 5.00 6.00 7.00 8.00 8.97 9.92 9.00 10.00 10.77 11.11 11.00 12.00 13.00 5.43 15.00 16.00 17.00 1: TOF MS ES+ 277.09 0.0500Da 362 16.00 17.00 1: TOF MS ES+ TIC 2.40e4 acetonitrile XIC clenbuterol % 100 14.00 8.74 0 1.00 X1_140422_SALTO_006 2.00 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 10.71 100 10.77 9.93 % 0.03 11.11 10.63 6.71 7.63 8.01 8.08 7.16 7.30 6.58 8.97 9.19 9.76 8.47 11.53 12.65 12.84 15.00 methanol TIC 15.05 15.17 14.95 12.90 13.67 14.07 15.80 16.35 2.69 0 1.00 X1_140422_SALTO_003 2.00 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 5.77 100 5.44 5.95 % 8.23 6.31 0.69 1.36 1.63 2.02 2.69 3.35 acetonitrile TIC 7.47 4.98 6.596.91 4.18 8.02 8.79 9.16 17.00 1: TOF MS ES+ TIC 1.80e4 15.16 15.37 15.60 9.62 10.00 10.49 10.66 14.93 0 Time 1.00 2.00 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 Figure 13: TIC and XIC chromatograms of a saliva sample containing a mixture of toxic compounds. Comparison of eluent with acetonitrile or methanol. Bottom chromatograms: TIC of acetonitrile and methanol. Middle chromatograms: XIC of clenbuterol using acetonitrile and methanol. Top chromatogram: XIC of salmeterol using acetonitrile and methanol. The chromatography was performed using a gradient. Table 13 shows the gradient parameters. Solvent A consists of 1% acetonitrile in water (v/v) containing 0.1% formic acid (v/v). Solvent B consists of 1% water in acetonitrile (v/v) containing 0.1% formic acid (v/v). Table 13: Gradient of the microUPLC system. Minutes Flow (μl/min) %A %B 0 3 99 1 1 3 99 1 4 3 60 40 10 3 0 100 10.5 3 99 1 15 3 99 1 25 Default flow rates on the microfluidics system go from 0.2 till 5 microliters per minute, depending on the column which is used, the choice of solvents and the sample injection volume. In this study a flow rate of 3 microliter per minute was used. The sample volume to be injected depends on different factors. First the amount of sample volume which is available. When there is a small amount available one has to be aware of the fact that most injection systems use more sample than the sample volume which is injected. Furthermore, usually one does not want to finish the whole sample volume, so the injection volume has to be chosen in such a way that there is enough sample left for a second injection. In this study a sample injection volume of 2 microliter was used. The repeatability of the iKey was tested by multiple injections of the same standard solution. Appendix VI shows overlaid chromatograms of d6-clenbuterol of 7 saliva samples within one validation day. It can be seen that the retention time and intensities of the peaks are similar, so the repeatability of the iKey is good. Also the pressure profile (the pressure of the system during the injection cycle) gives an indication of the stability and repeatability of the analytical column or in this case the iKey. Appendix VI shows the pressure profile of all saliva samples of validation day 2 (n=21). The profiles are overlapping well, which gives an indication of the correct functioning of the iKey. Also in Appendix VI a pressure profile can be seen of three saliva samples injected on three different validation days. Also here the profiles are overlapping well, which gives an indication of the correct functioning of the iKey over different days. The variation between different iKeys was tested by injecting the same sample at two different iKey’s. Figure 14 shows a TIC and a XIC of clenbuterol in a saliva sample spiked with a mixture of veterinary drugs injected on two different (used) iKey’s. It can be seen that the shape of the chromatogram (TIC) is very different and also the retention time of clenbuterol is significantly different (0.5 minutes difference). This experiment shows that one has to be critical when using multiple iKey’s, because chromatographic characteristics (RT, peak shape) can be different between them. This can be caused by the same factors compared to conventional LC columns; misuse of the iKey, type of eluent used on the iKey, type of sample injected on the iKey or age of the iKey. It should be mentioned that when using two (new or used) iKeys which are both in good shape the performance (RT, peak shape) is identical (no data shown, but can be provided on request). 26 X1_140723_SALTO_004 1: TOF MS ES+ 277.087 0.0500Da 2.72e3 5.31 100 iKey 2 XIC clenbuterol % 5.39 5.43 0 1.00 2.00 X1_140723_SALTO_003 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 4.71 4.74 100 18.00 1: TOF MS ES+ 277.087 0.0500Da 1.84e3 % iKey 1 XIC clenbuterol 1.60 0 1.00 2.00 X1_140723_SALTO_004 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 5.95 100 18.00 1: TOF MS ES+ TIC 9.59e4 iKey 2 TIC 5.31 6.45 % 5.18 8.07 5.43 9.97 8.468.53 9.27 4.59 10.57 14.85 14.13 13.45 9.60 1.36 11.06 12.03 12.26 13.23 14.59 15.10 15.33 0 1.00 2.00 X1_140723_SALTO_003 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 5.35 100 4.71 1.59 5.27 iKey 1 TIC 5.78 14.89 % 18.00 1: TOF MS ES+ TIC 7.52e4 15.04 10.63 7.43 1.51 15.28 10.77 11.45 0 Time 1.00 2.00 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 18.00 Figure 14: Comparison of two different iKey’s. TIC and XIC of clenbuterol of a spiked saliva sample. Shape of the chromatogram (TIC) as well as retention time of clenbuterol (XIC) are different between the iKey’s. MS: Before every MS analysis the correct working of the lock-spray have to be tested. This can be done manually by switching the lock spray to the MS inlet. When a convenient signal (user specified) is produced regarding intensity and stability, the lock spray is working in a correct way. Figure 15 shows a lock spray spectrum using a Leucine and Enkypheline mixture of 2000 μg/l. The masses with a star are the lock spray masses which should be detected. 27 20; blank saliva (T) + 1 ppm X1_141222_SALTO_val day 3_024 18 (5.676) * 397 100 3: TOF MS ES+ 3.68e3 * 556 % * 425 594 * 278 557 ! 279 398 85 59 86 120 136 163 177 426 205 336 221 79 90 !;103 0 50 100 137 150 199 262 ! 280 234 261 317 250 300 200 331 337 595 538 481 375 393 399 373 350 400 510 558 443 596 511 ! 493 465 512 450 500 ! ! 616 636 658 692 698 714 749 778 792 810 550 600 650 700 750 800 865 872 850 906 900 910 ! ! ! ! 937 958 996 986 950 m/z 1000 Figure 15: Lock spray spectrum of Leucine/Enkypheline mixture. The masses with a star are the lock spray masses which should be detected. Also before every MS analysis the MS should be calibrated (see 2.5). An example of a calibration file is shown in Appendix II. In UNIFI a targeted data processing method was build containing all compounds which are present in the standard mixtures (see chapter 3.1). The compound characteristics that were included in the library were retention time, monoisotopic mass and (if known) the fragment masses. The retention times of the compounds were determined by manually evaluation of the peaks in chromatograms for analyzed standard solutions containing some groups of compounds. The final compound list including retention times and monoisotopic masses can be found in appendix III (only validated compounds, see chapter 4.4). The compounds together with the retention times and monoisotopic masses were imported in a targeted processing method in UNIFI (see paragraph 2.6). Part of the processing method is included in appendix IV. Figure 16 to 18 show the results for a saliva sample fortified with 0.1 mg/l clenbuterol. Figure 16 shows the extracted ion chromatogram where clenbuterol can be found at retention time 5.12. In figure 17 a part of a low energy and a high energy spectrum of this clenbuterol peak can be seen. The peaks of clenbuterol, with experimental mass 277.07869 Da and its chlorine isotope peak 279.08188 (clenbuterol contains two chlorine atoms), are significantly higher in the low energy spectrum compared to the high energy spectrum. This is explained by the fact that with high energy part of the precursor ion is fragmented. Figure 18 shows the full spectra (high and low energy), where the fragment masses are identified. Figures 19 and 20 show a comparable chromatogram and high and low energy spectra for the toxin atropine in fortified saliva sample (0.1 mg/l). Also here the fragment ions are identified. In this way all compounds are identified based on the retention time, monoisotopic mass and fragment ions (if present in the method). If the deviation in retention time and the monoisotopic mass are within the set criteria (RT 0.1 minute and mass <25mDa, see paragraph 2.6) the compounds are identified. When there are also identified fragment ions, the identification is even more reliable. 28 Figure 16: Chromatogram of clenbuterol in saliva (saliva sample fortified with 0.1 mg/l clenbuterol). Figure 17: Part of the spectrum of [M+H]+ ion of clenbuterol (277.0787). Top: low energy scan. Bottom: high energy scan. The low energy scan shows high intensities of the peaks, the high energy scan low intensities. 29 Figure 18: Full spectrum of precursor and product ions of clenbuterol (277.0787). Top: low energy scan. Bottom: high energy scan. The low energy scan only shows the [M+H]+ ion of clenbuterol, the high energy scan shows the fragment ions of clenbuterol. 30 Figure 19: Chromatogram of atropine in saliva (saliva sample fortified with 0.1 mg/l atropine). Figure 20: Full spectrum of precursor and product ions of Atropine (290.17504). Top: low energy scan. Bottom: high energy scan. The low energy scan only shows the [M+H]+ ion of atropine, the high energy scan shows the fragment ions of atropine. 31 4.4 Validation Sample pre-treatment of the validation samples was performed according the method described in Appendix IX after all sample pre-treatment and LC-MS parameters were optimized. The validation itself was performed according an in-house validation protocol which is described in paragraph 2.7 and 3.3. This means that the validation was performed during 3 days with 20 different saliva samples. The acquisition data was processed using the processing method in UNIFI. Compounds are considered to be identified when UNIFI identifies a peak in a chromatogram and when this is visually confirmed as an actual peak by the reviewer. This means that a peak has to fulfill the criteria of retention time and mass deviation (see paragraph 2.6) and an additional check is performed. After data processing of validation day 1 it became clear the data was not suitable for further evaluation. Something went wrong in the LC-MS analysis causing retention time shifts of the chromatographic peaks. Due to this the samples of validation day 1 were analyzed again after being stored in the freezer for 1 week (see also stability of the samples extracts). After data reviewing it became clear that the reanalyzed data of validation day 1 could be used for the validation. Appendix X shows 2 examples of processed validation data of validation day 3. It can be seen that both compounds (trifloxystrobin and clenbuterol) were identified in all spiked saliva samples (n=7). Trifloxystrobin was unidentified in all blank saliva samples (selectivity samples) and in three of the 7 blank saliva samples clenbuterol was identified. Nevertheless, with a visual check clenbuterol could be considered as unidentified in these three samples, because the intensities of the peaks were to low (the integrated peak was not a real chromatographic peak). To prevent the software to generate false positive samples (falsely identified peaks) a peak intensity detection threshold could be included in the method. This was not done in this validation to prevent false negative (falsely unidentified peaks) samples to occur. CCß is the concentration value for which in 95% of the fortified samples the compound is identified. In this validation study 21 blank saliva samples were used. One sample (sample G, validation day 1) was considered to be an outlier, because the retention times of most compounds were not stable and did not pass the requirements. Probably something went wrong in the LC-MS analysis, which caused this problem. For every compound 19 of the 20 saliva samples have to give a positive identification at the validation level. Because 20 screen positive control samples have been used in this validation, the CCß is set to the target concentration (See, Guidelines for the validation of screening methods for residues of veterinary medicines,(33)). Appendix VII shows the number of identifications per sample at the two concentration levels. When no or less than 19 identification at 1 mg/l were found while at 0.1 mg/l 19 or 20 identifications were found this is due to an overloading problem of the TOF MS. Because the detector was overloaded the mass assignment was not accurate anymore. When this was the case the CCß of the compound was set to 0.1 mg/l. The table in Appendix VIII gives the CCß values (0.1 mg/l or 1 mg/l) for the identified compounds, which are in total 202 compounds. Compounds which did not succeed these requirements (so less than 19 identifications) were considered to be non-validated compound and were left out the results of this validation study. These were not included in in the table in Appendix VII and VIII. These were in total 94 compounds which corresponds with 31% of the total compounds in the tox mixes. One of the reasons some compounds were not identified lies in the problem that the more polar compounds were not retained well on the iKey. This has to do with the characteristics of the iKey. A solution to avoid this problem can be the use of a trapping column in front of the iKey. 32 For the selectivity the same 21 saliva samples were analyzed and processed with the same method as the fortified samples. Also in this case the outlier sample was left out. When a well-defined peak was detected at the retention time of a compound in one of the samples the method could be considered as not selective for this compound. For twelve compounds peaks were found (identified after automatic data processing and manual check) in the blank samples. This means the analysis is not selective for these compounds. The non-selective compounds can be found in table 14, between brackets are the number of samples were the compound was identified in the blank saliva samples (n=20). For all other compounds no peaks were founds in the blank samples, so the analysis is selective for these compounds. Table 14: Non-selective compounds from the validation experiments. Between brackets the number of samples (out of 20) which show no interfering peaks. Toxins # Veterinary drugs # Pesticides # coumarin (8/20) broomchloorbuterol (13/20) carbetamide (13/20) digitoxigenin (10/20) carbuterol (15/20) disulfoton-sulfone (18/20) santonin (13/20) ractopamine (9/20) fenbuconazole (16/20) pirimicarb (14/20) tebufenozide (13/20) trinexapac-ethyl (8/20) The ruggedness was performed according table 5, chapter 2.7 for all validated compounds. For all ruggedness samples all compounds were identified. This means the method is robust for the tested variations in the method for the validated compounds. From this it can be concluded that the addition of DMSO to prevent the compounds from not re-dissolving after evaporating has no added value. In case of a continuation of the project or an additional validation this should be taken into account. To determine the stability of the sample extracts the samples of validation day 1 were stored in the freezer for one week and analyzed again. Because the initial analysis of validation day 1 failed, the results of the stability test (samples of day 1 stored in the freezer for 1 week and re-analyzed) were used as a full validation day (validation day 1). Because of this it can be stated that the identified compounds mentioned in appendix VII are stable for one week in the freezer. The stability of the standard solutions has not been tested in this validation, because information on these stabilities was already obtained in other studies performed at RIKILT. Data on this is available in multiple RIKILT SOP’s. 33 5 Conclusion A complete method for the toxicological screening of bovine saliva was successfully developed and validated as part of this research project. This method includes a saliva sampling method, sample pretreatment, LC-MS analysis and data processing. A saliva sampling method was developed. Sampling can be performed using two different sampling devices depending on the amount of saliva needed. For amounts lower than 300 μl a swab has to be used while for amounts higher than 300 μl a cotton rope has to be used. After sampling the saliva has to be extracted from the devices or the devices have to be stored in the freezer as soon as possible. The main reason for sample pre-treatment is to get rid of proteins which are present in the saliva. To achieve this a protein precipitation method using acetonitrile was developed and tested. Three main compound groups were included in the study, namely pesticides, natural toxins and veterinary drugs. These compounds can be toxic for animals due to accidentally intake during feeding or negligence of the owner of the animal. They can also be administered on purpose like poisoning or overdosing. A microLC-microfluidics-TOF-MS method was developed for the identification of the compounds in saliva. Compared to conventional LC, with microLC a relatively low flow is used, 3 μl/min. An injection volume of 2 μl has been used. The microfluidics system consists of a Waters IonKey system, which includes an analytical column in a chip device (iKey), using ESI as an ionization technique. The IonKey source is placed in front of a QTOF. The acquisition method used is a so called MSe method were a full scan and a fragmentation scan are acquired alternately. In this way next to full scan data there is also fragmentation scan data available which can be used for more reliable identification of the compounds. A targeted library was developed using UNIFI software. The library was constructed using experimental obtained data using standard solutions containing pesticides, natural toxins and veterinary drugs. The library includes retention times, monoisotopic masses and if present fragment ions. The developed method was validated according 2002/657/EC to test the characteristics of the method. For a screening method the limit of detection (CCß), selectivity and ruggedness of the method and stability of the compounds and sample extracts needs to be validated. The validation was performed on three days with 20 different blank saliva samples. The samples were spikes at two levels, 0.1 and 1 mg/liter. These corresponds with the CCß levels of the validated compounds. The validation results show that with the method 203 compounds can be detected in saliva with a CCß of 0.1 or 1 mg/l (appendix VIII). The method was not selective for twelve compounds, interfering peaks were found in the chromatograms. The method was rugged for the tested variations on the sample pre-treatment and sample extracts are stable for one week in the freezer. Summarizing, within this research project a screening method for more than 200 toxic compounds was successfully developed. The method consists of a sampling procedure, sample pre-treatment and analyzing method using microLC-microfluidics-TOF-MS. Additionally, the method was validated according European legislation. 34 6 Recommendations and future perspectives Additional to the conclusion of this thesis some recommendation and future perspectives can be made. In this study only bovine saliva was validated. It would be useful to perform an additional validation with saliva from other animals as well. For example horses and dogs will be of great interest because owners of these animals have the tendency of being concerned about their animal, so most forensic saliva samples will be coming from these animals. Also instead of saliva, serum could be validated additionally. As already mentioned in this thesis, serum is a common matrix in forensic science, but not always easy to sample. In addition to the compounds which are validated in this study, it would be useful to extend the number of compounds. This can be done by adding new compound groups, such as human drugs, illicit drugs or cleaning agents. Another way of extending the amount of compounds is to improve the analysis method. With the used iKey it is not possible to analyze in the negative electrospray ionization mode. When the manufacturer develops a system where this is possible also compounds could be measured which have to be analyzed in negative ionization mode. To test the analytical system used, microfluidics systems of other manufacturers could be used. In the method development and validation it was found that more polar compounds could not be separated with the iKey, they eluted in the death volume of the column. This can be (partly) solved by using a trap column in front of the iKey. 35 7 References 1. Berny P, Caloni F, Croubels S, Sachana M, Vandenbroucke V, Davanzo F, et al. Animal poisoning in Europe. Part 2: Companion animals. The Veterinary Journal. 2010;183(3):255-9. Guitart R, Croubels S, Caloni F, Sachana M, Davanzo F, Vandenbroucke V, et al. Animal poisoning in Europe. Part 1: Farm livestock and poultry. The Veterinary Journal. 2010;183(3):249-54. Vudathala DK, Cummings MR, Murphy LA. A multiresidue screen for the analysis of toxicants in bovine rumen contents. Journal of Veterinary Diagnostic Investigation. 2014;26(4):531-7. Langel K, Gjerde H, Favretto D, Lillsunde P, Øiestad EL, Ferrara SD, et al. Comparison of drug concentrations between whole blood and oral fluid. Drug Testing and Analysis. 2014;6(5):461-71. Aps JKM, Martens LC. Review: The physiology of saliva and transfer of drugs into saliva. Forensic Science International. 2005;150(2–3):119-31. Cortinovis C, Caloni F. Epidemiology of intoxication of domestic animals by plants in Europe. Veterinary Journal. 2013;197(2):163-8. Vandenbroucke V, Van Pelt H, De Backer P, Croubels S. Animal poisonings in Belgium: A review of the past decade. Vlaams Diergeneeskundig Tijdschrift. 2010;79(4):259-68. Jansen SA, Kleerekooper I, Hofman ZL, Kappen IF, Stary-Weinzinger A, van der Heyden MA. Grayanotoxin poisoning: 'mad honey disease' and beyond. Cardiovascular toxicology. 2012;12(3):20815. Acute Vergiftigingen Bij Mens En Dier, jaaroverzicht 2008-2013. In: (2009-2014) ebNVIC, editor.: Rijksinstituut voor Volksgezondheid en Milieu (RIVM); 2009-2014. Anadón A, Martínez-Larrañaga MR, Castellano V. Chapter 78 - Poisonous plants of Europe. In: Gupta RC, editor. Veterinary Toxicology (Second Edition). Boston: Academic Press; 2012. p. 1080-94. K. Baert SC, N. Steurbaut, S. De Boever, G. Vercauteren, R. Ducatelle, A. Verbeken, P. De Backer. Two unusual cases of plant intoxication in small ruminants. Vlaams Diergeneeskundig Tijdschrift. 2005;74:149-53. Herfsttijloos gevonden in paardenhooi [press release]. Gezondheidsdienst voor Dieren2014. Kaufman E, Lamster IB. The Diagnostic Applications of Saliva— A Review. Critical Reviews in Oral Biology & Medicine. 2002;13(2):197-212. Fucci N, De Giovanni N, Chiarotti M. Simultaneous detection of some drugs of abuse in saliva samples by SPME technique. Forensic Science International. 2003;134(1):40-5. Kidwell DA, Holland JC, Athanaselis S. Testing for drugs of abuse in saliva and sweat. Journal of Chromatography B: Biomedical Sciences and Applications. 1998;713(1):111-35. Gao W, Stalder T, Kirschbaum C. Quantitative analysis of estradiol and six other steroid hormones in human saliva using a high throughput liquid chromatography-tandem mass spectrometry assay. Talanta. 2015;143:353-8. Cuevas-Córdoba B, Santiago-García J. Saliva: A fluid of study for OMICS. OMICS A Journal of Integrative Biology. 2014;18(2):87-97. Lamy E, Mau M. Saliva proteomics as an emerging, non-invasive tool to study livestock physiology, nutrition and diseases. Journal of proteomics. 2012;75(14):4251-8. Mandel ID, Wotman S. The salivary secretions in health and disease. ORAL SCIREV. 1976;Vol.8:25-47. Townsend S, Fanning L, O'Kennedy R. Salivary Analysis of Drugs—Potential and Difficulties. Analytical Letters. 2008;41(6):925-48. Haeckel RaH, P. Application of Saliva for Drug Monitoring. An In Vivo Model for Transmembrane Transport. Eur J Clin Chem Clin Biochem. 1996;34:171-91. Chensheng Lu Leigh CARAF. Determination of atrazine levels in whole saliva and plasma in rats: Potential of salivary monitoring for occupational exposure. Journal of Toxicology and Environmental Health. 1997;50(2):101-12. Borzelleca JF, Skalsky HL. The excretion of pesticides in saliva and its value in assessing exposure. Journal of Environmental Science and Health, Part B. 1980;15(6):843-66. Churchwell MI, Twaddle NC, Meeker LR, Doerge DR. Improving LC–MS sensitivity through increases in chromatographic performance: Comparisons of UPLC–ES/MS/MS to HPLC–ES/MS/MS. Journal of Chromatography B. 2005;825(2):134-43. Grumbach ES. Transferring yesterdays methods to tomorrows technology: the evolution from HPLC to UPLC. PITTCONN2005. Uliyanchenko E, van der Wal S, Schoenmakers PJ. Challenges in polymer analysis by liquid chromatography. Polymer Chemistry. 2012;3(9):2313-35. Sneekes ER, L. Swart, R. Nano LC: Principles, Evolution, and State-of-the-Art of the Technique2011; 29(10). Gerssen AB, M.H. et al. The use of Trizaic NanoTile Technology for steroid analysis. RIKILT report. 2012. Hoffmann de E, Stroobant V. Mass spectrometry: principles and application. Third edition ed: Wiley; 2007. Commission Decision 2002/657, (2002). Stolker AAM. Identification of residues by LC-MS. The application of new EU guidelines. Analusis. 2000. Schipper R, Loof A, de Groot J, Harthoorn L, Dransfield E, van Heerde W. SELDI-TOF-MS of saliva: Methodology and pre-treatment effects. Journal of Chromatography B. 2007;847(1):45-53. Guidelines for the validation of screening methods for residues of veterinary medicines, (2010). 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. 16. 17. 18. 19. 20. 21. 22. 23. 24. 25. 26. 27. 28. 29. 30. 31. 32. 33. 36 Appendix I: Camera view of IonKey source interior Baffle in ESI position Baffle in Lock spray position 37 Appendix II: TOF-MS calibration file 38 Appendix III: Compound list with RT and monoisotopic mass Toxins Aconitine Aflatoxin B1 Aflatoxin B2 Aflatoxin G1 Ajmalicine Aristolochia Acid II Atropine Colchicine Convallatoxin Coumarin Digitoxigenin Digoxin Erucifoline-N-Oxide Genistein Heliotrine Imperatorin Jacobine Kavain Lobeline Narceine Noscapine Ochratoxin A Ouabain (Strophanthin G-) Papaverine Podophyllotoxin Rescinnamine Reserpine Retorsine Santonin Senecionine Senecionine-N-Oxide Seneciphylline Senkirkine Solanine Alpha Umbelliferone Vincamine RT 7.53 6.94 6.65 661 5.98 6.61 4.60 6.59 6.39 5.49 7.77 7.40 5.41 8.98 4.46 8.98 5.10 8.09 6.59 6.15 6.00 8.88 4.98 5.92 9.46 8.08 7.88 5.12 7.08 5.01 5.12 4.55 5.38 6.80 4.70 5.70 Mass Veterinary drugs 645.3149 Broombuterol 312.0634 Broomchloorbuterol 314.0790 Carbuterol 328.0583 Cimaterol 352.1787 Clenbuterol 311.0430 Clencyclohexerol 289.1678 Clenpenterol 399.1682 Clenproperol 550.2778 Demecloxycline 146.0368 Doxycycline 374.2457 Emamectin 780.4296 Eprinomectine 365.1475 Hydroxymethylclenbuterol 270.0528 Isoxsuprine 313.1889 Mabuterol 270.0892 Mapenterol 351.1682 Monensin 230.0943 Moxidectine 337.2042 Oxytetracycline 445.1737 Ractopamine 413.1475 Salbutamol 403.0823 Salmeterol 584.2833 Tetracycline 339.1471 Tulobuterol 414.1315 634.2890 608.2734 351.1682 246.1256 335.1733 351.1682 333.1576 365.1838 867.4980 162.0317 354.1943 RT 5.31 5.36 1.60 1.59 5.12 4.05 5.75 4.58 5.07 6.00 9.58 10.64 4.47 5.69 5.75 6.25 11.36 11.23 4.60 4.88 1.65 7.90 4.81 5.15 Mass 363.9786 320.0291 267.1583 219.1372 276.0796 318.0902 290.0953 262.0640 464.0986 444.1533 885.5238 913.5188 292.0745 301.1678 310.1060 324.1216 670.4292 639.3771 460.1482 301.1678 239.1521 415.2723 444.1533 227.1077 39 Appendix III (continued): Compound list with RT and monoisotopic mass Pesticides Acetamiprid Aclonifen Aldicarb Sulfon Aldicarb Sulfoxide Ametoctradin Atrazine Azadirachtin Azoxystrobin Benzoylprop-Ethyl Bitertanol Bixafen Bupirimate Buprofezin Carbaryl Carbetamide Carbofuran Carfentrazone-Ethyl Chlorantraniliprole Chlorbromuron Chloridazon Chlorpyrifos Clomazone Clothianidin Cyazofamid Cybutryne Cyproconazole I Cyproconazole II Cyprodinil Cythioate Diazinon Dichlorvos Difenoconazole I Difenoconazole II Diflufenican Dimethenamid Dimethoate RT 5.53 8.92 7.22 7.70 8.72 7.37 9.26 8.88 9.90 9.33 9,49 8.30 9.06 7.33 6.33 7.03 9.60 8.36 8.66 5.04 10.61 8.17 4.89 9.60 7.65 8.63 8.63 7.81 6.95 9.77 6.65 9.72 9.72 10.17 8.61 5.09 Mass 222.0672 264.0302 222.0674 206.0725 275.2110 215.0938 720.2629 403.1168 365.0585 337.1790 413.0310 316.1569 305.1562 201.0790 236.1161 221.1052 411.0364 480.9708 291.9614 221.0356 348.9263 239.0713 249.0087 324.0448 253.1361 291.1138 291.1138 225.1266 296.9895 304.1010 219.9459 405.0647 405.0647 394.0741 275.0747 228.9996 Pesticides Dimethomorph I Dimethomorph II Dimoxystrobin Disulfoton Disulfoton-Sulfone Diuron Dodemorph Ethirimol Ethoprophos Etoxazole Fenamidone Fenamiphos Fenbuconazole Fenhexamid Fenobucarb Fenpropimorph Flamprop-Isopropyl Flamprop-Methyl Florasulam Fluazifop Fluazifop-P-Butyl Flufenacet Fluopicolide Fluoxastrobin Flusilazole Flutolanil Flutriafol Foramsulfuron Fosthiazate Haloxyfop Haloxyfop-P-Methyl Hexaconazole Imazalil Imidacloprid Iprodione Iprovalicarb RT 8.47 8.61 9.37 6.10 8.33 7.67 7.81 5.20 8.91 10.01 8.87 8.91 9.25 9.01 8.47 8.09 9.87 9.15 7.15 9.50 10.58 9.34 9.06 9.41 9.20 9.28 7.71 7.07 7.55 10.11 10.17 9.25 7.20 5.14 8.94 8.83 Mass 387.1237 387.1237 326.1630 274.0285 306.0183 232.0170 281.2719 209.1528 242.0564 359.1697 311.1092 303.1058 336.1142 301.0636 207.1259 303.2562 363.1037 335.0724 359.0300 327.0718 383.1344 363.0665 381.9654 458.0793 315.1003 323.1133 301.1027 452.1114 283.0466 361.0329 375.0485 313.0749 296.0483 255.0523 329.0334 320.2100 40 Appendix III (continued): Compound list with RT and monoisotopic mass Pesticides Isoprocarb Isoproturon Isopyrazam Isoxaben Kresoxim-Methyl Lenacil Linuron Mandipropamid Mecoprop Mepanipyrim Mepronil Mesosulfuron-Methyl Metalaxyl Metamitron Metazachlor Metconazole Methidathion Methoxyfenozide Metolachlor Metoxuron Metrafenone Metribuzin Mevinphos Myclobutanil Nicosulfuron Ouabain (Strophanthin G-) Oxadixyl Parathion Penconazole Pencycuron Picoxystrobin Pirimicarb Pirimiphos-Methyl Podophyllotoxin Prochloraz Profenofos RT 8.02 7.71 10.01 9.13 9.62 7.04 8.50 9.06 6.46 8.87 9.10 7.95 7.82 4.70 8.02 9.32 8.40 9.16 9.18 6.22 10.15 6.48 5.24 8.89 6.75 4.98 6.73 9.72 9.21 10.00 9.73 5.13 9.56 9.46 8.66 10.16 Mass 193.1103 206.1419 359.1809 332.1736 313.1314 234.1368 248.0119 411.1237 214.0397 223.1109 269.1416 503.0781 279.1471 202.0855 277.0982 319.1451 301.9619 368.2100 283.1339 228.0666 408.0572 214.0888 224.0450 288.1142 410.1009 584.2833 278.1267 291.0330 283.0643 328.1342 367.1031 238.1430 305.0963 414.1315 375.0308 371.9351 Pesticides Propiconazole I Propiconazole II Propoxur Propyzamide Prosulfocarb Pyraclostrobin Pyroxsulfam Quinmerac Quinoclamine Quinoxyfen Quizalofop-Ethyl Silthiofam Simazine Spinosyn-A Spiroxamine I Spiroxamine II Sulcotrione Tebuconazole Tebufenozide Tebufenpyrad Terbucarb Terbuthylazine Terbutryn Tetraconazole Thiacloprid Thiophanate-Methyl Topramezone Triazophos Triazoxide Tricyclazole Trifloxystrobin Trinexapac-Ethyl Triticonazole Tritosulfuron Vamidothion Zoxamide RT 9.39 9.39 6.94 8.85 9.18 9.90 7.34 7.22 6.41 10.06 10.24 9.45 6.42 9.04 8.10 8.10 7.30 9.12 9.55 10.26 10.06 8.44 7.41 9.13 6.09 6.96 6.60 9.25 5.88 5.75 10.31 7.95 8.69 9.70 5.26 9.78 Mass 341.0698 341.0698 209.1052 255.0218 251.1344 387.0986 434.0620 221.0244 207.0087 306.9967 372.0877 267.1113 201.0781 731.4608 297.2668 297.2668 328.0172 307.1451 352.2151 333.1608 277.2042 229.1094 241.1361 371.0215 252.0236 342.0456 363.0889 313.0650 247.0261 189.0361 408.1297 252.0998 317.1295 445.0279 287.0415 335.0247 41 Appendix IV: Processing method (partly) 42 Appendix V: Experimental scheme of validation day 3 sample # 1 2 3 4 5 6 7 sample description blank saliva (O) blank saliva (P) blank saliva (Q) blank saliva (R) blank saliva (S) blank saliva (T) blank saliva (U) volume µl 100 100 100 100 100 100 100 VL mg/l 0 0 0 0 0 0 0 spike solution (ng/µl) 1 10 8 9 10 11 12 13 14 blank saliva (O) + 0.1 ppm blank saliva (P) + 0.1 ppm blank saliva (Q) + 0.1 ppm blank saliva (R) + 0.1 ppm blank saliva (S) + 0.1 ppm blank saliva (T) + 0.1 ppm blank saliva (U) + 0.1 ppm 100 100 100 100 100 100 100 0.1 0.1 0.1 0.1 0.1 0.1 0.1 15 16 17 18 19 20 21 blank saliva (O) + 1 ppm blank saliva (P) + 1 ppm blank saliva (Q) + 1 ppm blank saliva (R) + 1 ppm blank saliva (S) + 1 ppm blank saliva (T) + 1 ppm blank saliva (U) + 1 ppm 100 100 100 100 100 100 100 1 1 1 1 1 1 1 10 10 10 10 10 10 10 5 5 5 5 5 5 5 22 23 24 25 26 27 Robustness 1 Robustness 2 Robustness 3 Robustness 4 Robustness 5 Robustness 6 100 100 100 100 100 100 1 1 1 1 1 1 10 10 10 10 10 10 5 5 5 5 5 5 28 29 30 chem chem + IS chem + IS + 1 ppm 10 5 5 10 10 10 10 10 10 10 IS 13C-Caf/clen-d6 (10 ng/µl) 5 5 5 5 5 5 5 5 5 5 5 5 5 5 43 Appendix VI: iKey repeatability Figure 6: Overlaid pressure profiles of 21 saliva samples within one day (validation day 2) Figure 7: Overlaid chromatogram (IS, clenbuterol-d6) of 7 saliva samples within one day (validation day 2) 44 Appendix VI (continued): iKey repeatability 1; blank saliva (O) X1_141222_SALTO_val day 3_003 4800.000 1.12 nBSM System Pressure Range: 2785 6.23 15.39 4600.000 4400.000 4200.000 4000.000 3800.000 3600.000 3400.000 3200.000 3000.000 2800.000 psi 2600.000 2400.000 2200.000 2000.000 1800.000 1600.000 1400.000 1200.000 1000.000 800.000 600.000 400.000 200.000 0.000 Time 1.00 2.00 3.00 4.00 5.00 6.00 7.00 8.00 9.00 10.00 11.00 12.00 13.00 14.00 15.00 16.00 17.00 18.00 Figure 8: Overlaid pressure profiles of 3 saliva samples between days (validation day 1, 2 and 3) 45 Appendix VII: Validated compounds with detection results Number of samples in which the compound was identified (out of 20 validation samples). *: less than 19 or no identifications Toxins Aconitine Aflatoxin B1 Aflatoxin B2 Aflatoxin G1 Ajmalicine Aristolochia Acid II Atropine Colchicine Convallatoxin Coumarin Digitoxigenin Digoxin Erucifoline-N-Oxide Genistein Heliotrine Imperatorin Jacobine Kavain Lobeline Narceine Noscapine Ochratoxin A Ouabain (Strophanthin G-) Papaverine Podophyllotoxin Rescinnamine Reserpine Retorsine Santonin Senecionine Senecionine-N-Oxide Seneciphylline Senkirkine Solanine Alpha Umbelliferone Vincamine 0.1 mg/l 19/20 * 20/20 * 20/20 * 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 19/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 * 20/20 1 mg/l * 20/20 20/20 19/20 20/20 20/20 19/20 18/20 20/20 20/20 20/20 20/20 20/20 20/20 * 20/20 20/20 20/20 19/20 * 19/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 17/20 20/20 * 20/20 20/20 20/20 Veterinary drugs Broombuterol Broomchloorbuterol Carbuterol Cimaterol Clenbuterol Clencyclohexerol Clenpenterol Clenproperol Demecloxycline Doxycycline Emamectin Eprinomectine Hydroxymethylclenbuterol Isoxsuprine Mabuterol Mapenterol Monensin Moxidectine Oxytetracycline Ractopamine Salbutamol Salmeterol Tetracycline Tulobuterol 0.1 mg/l * 20/20 20/20 19/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 19/20 20/20 20/20 20/20 20/20 * 20/20 20/20 20/20 * 20/20 20/20 20/20 1 mg/l 20/20 20/20 18/20 20/20 * 20/20 20/20 20/20 20/20 20/20 18/20 19/20 * 20/20 15/20 20/20 20/20 20/20 20/20 20/20 19/20 20/20 20/20 20/20 46 Appendix VII (continued): Validated compound with CCß values Number of samples in which the compound was identified (out of 20 validation samples). *: less than 19 or no identifications Pesticides Acetamiprid Aclonifen Aldicarb Sulfon Aldicarb Sulfoxide Ametoctradin Atrazine Azadirachtin Azoxystrobin Benzoylprop-Ethyl Bitertanol Bixafen Bupirimate Buprofezin Carbaryl Carbetamide Carbofuran Carfentrazone-Ethyl Chlorantraniliprole Chlorbromuron Chloridazon Chlorpyrifos Clomazone Clothianidin Cyazofamid Cybutryne Cyproconazole I Cyproconazole II Cyprodinil Cythioate Diazinon Dichlorvos Difenoconazole I Difenoconazole II Diflufenican Dimethenamid Dimethoate 0.1 mg/l 20/20 20/20 * * 20/20 20/20 * 20/20 20/20 19/20 20/20 20/20 20/20 * 20/20 20/20 * 20/20 20/20 20/20 * 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 19/20 20/20 20/20 16/20 20/20 20/20 1 mg/l 20/20 20/20 20/20 20/20 20/20 20/20 20/20 19/20 20/20 20/20 20/20 13/20 20/20 19/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 13/20 20/20 20/20 20/20 20/20 18/20 20/20 20/20 20/20 20/20 20/20 20/20 Pesticides Dimethomorph I Dimethomorph II Dimoxystrobin Disulfoton Disulfoton-Sulfone Diuron Dodemorph Ethirimol Ethoprophos Etoxazole Fenamidone Fenamiphos Fenbuconazole Fenhexamid Fenobucarb Fenpropimorph Flamprop-Isopropyl Flamprop-Methyl Florasulam Fluazifop Fluazifop-P-Butyl Flufenacet Fluopicolide Fluoxastrobin Flusilazole Flutolanil Flutriafol Foramsulfuron Fosthiazate Haloxyfop Haloxyfop-P-Methyl Hexaconazole Imazalil Imidacloprid Iprodione Iprovalicarb 0.1 mg/l 20/20 20/20 20/20 20/20 18/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 19/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 19/20 19/20 20/20 20/20 20/20 20/20 20/20 1 mg/l 20/20 20/20 20/20 20/20 19/20 20/20 20/20 20/20 20/20 20/20 15/20 20/20 20/20 20/20 20/20 19/20 20/20 19/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 17/20 20/20 20/20 19/20 20/20 13/20 20/20 20/20 20/20 * 47 Appendix VII (continued): Validated compound with CCß values Number of samples in which the compound was identified (out of 20 validation samples). *: less than 19 or no identifications Pesticides Isoprocarb Isoproturon Is opyrazam Isoxaben Kresoxim-Methyl Lenacil Linuron Mandipropamid Mecoprop Mepanipyrim Mepronil Mesosulfuron-Methyl Metalaxyl Metamitron Metazachlor Metconazole Methidathion Methoxyfenozide Metolachlor Metoxuron Metrafenone Metribuzin Mevinphos Myclobutanil Nicosulfuron Ouabain (Strophanthin G-) Oxadixyl Parathion Penconazole Pencycuron Picoxystrobin Pirimicarb Pirimiphos-Methyl Podophyllotoxin Prochloraz Profenofos 0.1 mg/l 20/20 20/20 20/20 20/20 * 20/20 20/20 20/20 19/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 * 1 mg/l 20/20 * 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 * 20/20 20/20 18/20 20/20 20/20 20/20 20/20 19/20 * 20/20 20/20 20/20 20/20 20/20 20/20 14/20 20/20 20/20 * * 20/20 20/20 20/20 Pesticides Propiconazole I Propiconazole II Propoxur Propyzamide Prosulfocarb Pyraclostrobin Pyroxsulfam Quinmerac Quinoclamine Quinoxyfen Quizalofop-Ethyl Silthiofam Simazine Spinosyn-A Spiroxamine I Spiroxamine II Sulcotrione Tebuconazole Tebufenozide Tebufenpyrad Terbucarb Terbuthylazine Terbutryn Tetraconazole Thiacloprid Thiophanate-Methyl Topramezone Triazophos Triazoxide Tricyclazole Trifloxystrobin Trinexapac-Ethyl Triticonazole Tritosulfuron Vamidothion Zoxamide 0.1 mg/l 20/20 20/20 19/20 20/20 * 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 19/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 20/20 19/20 19/20 20/20 20/20 20/20 20/20 * 20/20 19/20 20/20 20/20 1 mg/l 20/20 20/20 20/20 20/20 20/20 20/20 20/20 18/20 20/20 20/20 20/20 20/20 20/20 18/20 20/20 20/20 20/20 * 20/20 20/20 20/20 20/20 17/20 20/20 20/20 20/20 19/20 16/20 17/20 20/20 20/20 20/20 * 19/20 20/20 20/20 48 Appendix VIII: Validated compound with CCß values Toxins Aconitine Aflatoxin B1 Aflatoxin B2 Aflatoxin G1 Ajmalicine Aristolochia Acid II Atropine Colchicine Convallatoxin Coumarin Digitoxigenin Digoxin Erucifoline-N-Oxide Genistein Heliotrine Imperatorin Jacobine Kavain Lobeline Narceine Noscapine Ochratoxin A Ouabain (Strophanthin G-) Papaverine Podophyllotoxin Rescinnamine Reserpine Retorsine Santonin Senecionine Senecionine-N-Oxide Seneciphylline Senkirkine Solanine Alpha Umbelliferone Vincamine 0.1 mg/l x 1 mg/l x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x Veterinary drugs Broombuterol Broomchloorbuterol Carbuterol Cimaterol Clenbuterol Clencyclohexerol Clenpenterol Clenproperol Demecloxycline Doxycycline Emamectin Eprinomectine Hydroxymethylclenbuterol Isoxsuprine Mabuterol Mapenterol Monensin Moxidectine Oxytetracycline Ractopamine Salbutamol Salmeterol Tetracycline Tulobuterol 0.1 mg/l 1 mg/l x x x x x x x x x x x x x x x x x x x x x x x x x x 49 Appendix VIII (continued): Validated compound with CCß values Pesticides Acetamiprid Aclonifen Aldicarb Sulfon Aldicarb Sulfoxide Ametoctradin Atrazine Azadirachtin Azoxystrobin Benzoylprop-Ethyl Bitertanol Bixafen Bupirimate Buprofezin Carbaryl Carbetamide Carbofuran Carfentrazone-Ethyl Chlorantraniliprole Chlorbromuron Chloridazon Chlorpyrifos Clomazone Clothianidin Cyazofamid Cybutryne Cyproconazole I Cyproconazole II Cyprodinil Cythioate Diazinon Dichlorvos Difenoconazole I Difenoconazole II Diflufenican Dimethenamid Dimethoate 0.1 mg/l x x 1 mg/l x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x Pesticides Dimethomorph I Dimethomorph II Dimoxystrobin Disulfoton Disulfoton-Sulfone Diuron Dodemorph Ethirimol Ethoprophos Etoxazole Fenamidone Fenamiphos Fenbuconazole Fenhexamid Fenobucarb Fenpropimorph Flamprop-Isopropyl Flamprop-Methyl Florasulam Fluazifop Fluazifop-P-Butyl Flufenacet Fluopicolide Fluoxastrobin Flusilazole Flutolanil Flutriafol Foramsulfuron Fosthiazate Haloxyfop Haloxyfop-P-Methyl Hexaconazole Imazalil Imidacloprid Iprodione Iprovalicarb 0.1 mg/l x x x x 1 mg/l x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x 50 Appendix VIII (continued): Validated compound with CCß values Pesticides Isoprocarb Isoproturon Isopyrazam Isoxaben Kresoxim-Methyl Lenacil Linuron Mandipropamid Mecoprop Mepanipyrim Mepronil Mesosulfuron-Methyl Metalaxyl Metamitron Metazachlor Metconazole Methidathion Methoxyfenozide Metolachlor Metoxuron Metrafenone Metribuzin Mevinphos Myclobutanil Nicosulfuron Ouabain (Strophanthin G-) Oxadixyl Parathion Penconazole Pencycuron Picoxystrobin Pirimicarb Pirimiphos-Methyl Podophyllotoxin Prochloraz Profenofos 0.1 mg/l x x x x 1 mg/l x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x Pesticides Propiconazole I Propiconazole II Propoxur Propyzamide Prosulfocarb Pyraclostrobin Pyroxsulfam Quinmerac Quinoclamine Quinoxyfen Quizalofop-Ethyl Silthiofam Simazine Spinosyn-A Spiroxamine I Spiroxamine II Sulcotrione Tebuconazole Tebufenozide Tebufenpyrad Terbucarb Terbuthylazine Terbutryn Tetraconazole Thiacloprid Thiophanate-Methyl Topramezone Triazophos Triazoxide Tricyclazole Trifloxystrobin Trinexapac-Ethyl Triticonazole Tritosulfuron Vamidothion Zoxamide 0.1 mg/l x x x x 1 mg/l x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x x 51 Appendix IX: Final sample pre-treatment method 100 µl saliva in a 1.5 ml Eppendorf Tube Control samples Tox mix 1 mg/l (µl) Tox mix 10 mg/l (µl) Internal standard mix 10 mg/l (µl) - - 5 10 - 5 - 10 5 Control sample 0 mg/l Control sample 0.1 mg/l Control sample 1 mg/l Add 5 µl IS to the samples Add 250 µl acetonitrile to the samples Shake thoroughly with a vortex for 30 seconds Centrifuge 10 minutes at 10 000 g Transfer the supernatant into a 1.5 ml total recovery LC-MS vial Add 5 µl DMSO Evaporate the sample under a nitrogen gas flow at 40°C Reconstitute with 100 µl water and cap the vial. Vortex thoroughly for 30 seconds and place in an ultrasonic bath for 1 minute. 52 Appendix X: Results of validation day 3 53