user guide
advertisement

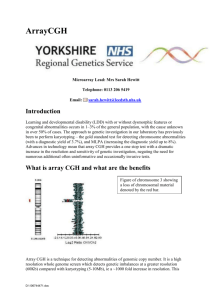
Prenatal Arrays user guide Microarray Comparative Genomic Hybridisation (array CGH) has been in service in the Leeds Cytogenetics lab since 2012 as a first line postnatal investigation for children with unexplained learning difficulties and/or dysmorphic features. The technology provides a high resolution genome-wide analysis of genomic imbalance and as a result the abnormality detection rate is twice that achieved by conventional cytogenetics. As chromosomal anomalies account for 20~25% of major fetal anomalies, array CGH is also increasingly being used to replace karyotyping for this referral group. There have been several recent studies on the application of array CGH in prenatal diagnosis which show that array CGH is more effective than karyotyping at detecting pathogenic genomic imbalance [refs PD Brady et al, 2014, B Oneda et al, 2014, and RJ Wapner et al, 2014, Vanakker et al 2014. In the UK, an EME-funded study (the EACH project) which ran between 2012 and 2014 was designed to evaluate the patient and professional views of the technology and the barriers/facilitators to its implementation into service. This study ended in May 2014 and is due to report the findings. The Leeds Cytogenetics lab participated in this study and has capitalised on its involvement to develop robust and reliable processes and testing pathways for prenatal arrays. It is anticipated that there will be a recommendation that prenatal arrays are introduced into diagnostic service for referrals with abnormal ultrasound findings. In the meantime, clear local guidelines have been produced detailing how the Leeds cytogenetic lab applies array CGH in a prenatal setting. This document is designed to provide current information about the local policy regarding prenatal arrays and information about the technology. The local policy is subject to amendment following publications and recommendations from the EACH study, and any recommendations set by the Royal College of Obstetricians and Gynaecologists and the Association for Clinical Genetic Science (ACGS) Quality Committee (for best practice guidelines). Users will be informed of any major changes in policy. Document1 What is array CGH and what are the benefits Figure of chromosome 8 showing a loss of chromosomal material denoted by the red bar Array CGH is a technique for detecting abnormalities of genomic copy number. It is a high resolution whole genome screen which detects genetic imbalances at a greater resolution (~60Kb) compared with karyotyping (5-10Mb), ie a ~1000 fold increase in resolution. This increase in resolution detects a higher number of genomic gains and losses (imbalances) than can be seen using QF-PCR & karyotype. Array CGH also accurately identifies the pathogenic genes involved in chromosome imbalances. This gives more accurate information on the cause of the fetus’s phenotype and enables better patient management. Who will be tested by array CGH? Prenatal array CGH will be introduced into our laboratory for selected cases for all referring centers on the 13th February 2015. All referrals will initially be tested by QF-PCR for sexing and rapid aneuploidy assessment for chromosomes X,Y,13,18 and 21. Where the QF-PCR result is normal prenatal arrays will be performed in the following cases unless stated on the referral form by the referring clinician: 1) 2) 3) 4) Document1 A pregnancy with an abnormal ultrasound scan A pregnancy with an increased NT of >3.5mm Other specific clinician requests A pregnancy in a family known to carry a clinically significant genomic imbalance detected pre- or postnatally by array CGH or another molecular method. Specimens required for QF-PCR, Array CGH and karyotype to be performed: Amniotic Fluid - Minimum 15-20ml CVS - 5 fronds Parental bloods will be requested by the laboratory if required Suboptimal Samples Where DNA quality from uncultured tissue is unsuitable for array CGH, it may be necessary to extract DNA from cultured cells. In such circumstances, the lab will notify the referring obstetrician of the need for performing array CGH on cultured cells as this will generate a delay in reporting time. Prenatal Pathway AF or CVS Serum screen risk or maternal age >3.5mm nuchal or other ab scan FH balanced chromosome abnormality QF-PCR 13,18,21,X,Y QF-PCR 13,18,21,X,Y QF-PCR 13,18,21,X,Y Normal Result Abnormal Result Normal Result Report Report Report Report Array CGH Culture Report Report Culture Document1 Report Normal Result Abnormal Result Turnaround Times (TAT) Our professional body the Association for Clinical Genetic Science (ACGS) issues best practice guidelines which define the recommended TAT for prenatal karyotyping as 95% of cases reported within 14 calendar days of sample receipt. At present it is not feasible to turn prenatal arrays around within this timeframe, although all efforts will be made by the lab to work towards achieving this. The lab will initially endeavour to report a prenatal array result within 21 days of sample receipt for a first line array, or within 28 days for an array performed after karyotyping due to an abnormal result. This will include any confirmatory studies necessary in the event of finding a pathogenic imbalance. Where it is necessary to analyse parental samples for confirmation/clarification purposes, an interim prenatal array CGH report will be issued and a final report when any necessary parental studies are completed. Where DNA quality from uncultured tissue is unsuitable for array CGH, it may be necessary to extract DNA from cultured cells. This will add on further delays to reporting times as DNA extraction can only take place when cultures are ready (about 14 days from receipt). Platform used Array CGH will be performed on extracted DNA using 8x60k ISCA design oligonucleotide arrays supplied through Illumina. The average resolution of the array is 60Kb. Only clinically significant imbalances will be reported. Interpretation and Reporting The purpose of a prenatal array in an ongoing pregnancy is to ascertain whether there is a genetic imbalance present that is clinically significant with respect to the features detected on antenatal scan or whether a familial imbalance is present in a fetus. Interpretation for ongoing pregnancies 1) Imbalances will only be reported if they are interpreted as likely to be clinically significant to the fetus in relation to the supplied USS features. Guidance will be taken from the locally held table produced by the EACH study. The guidelines were devised with multicenter input. Clinically significant findings will include recognised microdeletion and duplication syndromes, as well as other imbalances where the literature details association with significant problems. 2) Neurosusceptibility loci have variable expression and penetrance and therefore the phenotype of an individual with one of these imbalances may vary from within normal limits to developmental delay, autism and schizophrenia. As phenotypic certainty cannot be offered and in line with national consensus only a limited number of these loci that are associated with an increased incidence of anomalies detected on scan will be reported. Document1 3) Due to the increased resolution of the screening there may be occasional unexpected incidental findings. Incidental findings will be discussed with the obstetrician, Clinical Geneticists and laboratory staff on a case to case basis and will only be reported if they will have implications later for the individual or relative and by reporting them there will be a health benefit. 4) Benign imbalances or imbalances classified as variants of uncertain significance (either likely benign or uncertain) will not be reported and will not be validated. Interpretation for non-viable / terminated pregnancies If a pregnancy is no longer ongoing (after discussion with obstetric team/clinical geneticist) and on analysis an imbalance is detected that may either: 1) Have contributed to the phenotypic features of the fetus or the demise of the pregnancy, or 2) Be clinically significant for other family members (neurosusceptibility loci, incidental findings). These findings will be reported. Confirmation and follow-up If a clinically significant imbalance is detected further confirmation studies will be performed. These may take the form of FISH studies, Q-PCR (different to QF-PCR) studies, MLPA, karyotyping or other techniques as appropriate. Parental studies will also be undertaken to determine whether the imbalance has arisen de-novo or has been inherited. These studies may add extra days to the reporting time. Reports Following interpretation of the imbalances and adhering to the policy detailed above reports will take the following format: 1) No imbalances detected relevant to the fetus’s problems. NO CLINICALLY SIGNIFICANT IMBALANCE DETECTED Microarray analysis on DNA extracted from chorionic villi/amniotic fluid sample has shown a fe/male profile with no imbalances of known clinical significance detected. As such no further work will be carried out on this sample. Technical details: BlueGnome ISCA 8x60k OligoArray v2.0 array (chip number ******) and analysed with Bluefuse Multi software (v4.1, v2.0 gal, ensembl build 70, GRCh37). 2) Imbalance detected which can be associated with the scan findings Abnormal Report Document1 FINAL REPORT: FAMILIALLY INHERITED 5p DELETION AND 16p DUPLICATION DETECTED BY ARRAYCGH AND CONFIRMED BY G-BANDING Microarray analysis of DNA extracted from direct chronic villus material has shown a female profile with two large imbalances involving material derived from the short arm of chromosome 5 and the short arm of a chromosome 16. The first is a deletion of 5p15.33-5p15.33 which is estimated to be between a minimum of 2.1Mb (DNA positions:22179-2167668) and a maximum of 2.3Mb (DNA positions:1-2274717.5) in size and contains 26 genes including 5 OMIM morbid genes. The second imbalance detected is a duplication of 16p13.3-16p11.2 which is estimated to be 32Mb (minimum DNA positions:93748-32066997 and maximum DNA positions:1-32091684.5) in size. This region contains 428 genes including 55 OMIM morbid genes. This patient has been added to decipher showing the genes included in this region (see attached Decipher report - YRxxxxx). G-banded chromosome analysis has confirmed that these imbalances are the result of a maternally derived unbalanced translocation between the short arm of a chromosome 5 and the short arm of a chromosome 16, with the duplicated 16 material positioned on the deleted 5p (see CVxxxx and Bxxxx). It is highly likely that this unbalanced rearrangement is the cause of the scan findings seen in this foetus. Advice on prenatal diagnosis may be helpful in any future pregnancy involving xxx and her partner. This patient and her partner may benefit from genetic counselling which can be arranged at the Department of Clinical Genetics, Chapel Allerton, Leeds. Technical details: BlueGnome ISCA 8x60k OligoArray v2.0 array (chip number ******) and analysed with Bluefuse Multi software (v3.1, v2.0 gal, ensembl build 62, GRCh37). Results All array results will be telephoned to the obstetricians. Unusual abnormal array results will also be given to a Clinical Geneticist. Limits to Array CGH technique and reporting restrictions - Array CGH will not detect all genetic abnormalities - Array CGH cannot detect balanced chromosome rearrangements such as reciprocal translocations and inversions. Karyotyping will be carried out for patients suspected of these abnormalities. - Array CGH cannot detect low level mosaicism (< 30% mixture of abnormal and normal cells) - Anomalies may occur which cannot always be recognised or properly interpreted. - Not all imbalances will be reported (as detailed above and in the laboratories policy on prenatal arrays). Some unreported imbalances may become clinically significant postnatally. Frequently Asked Questions What if I can’t obtain parental blood samples? Document1 We are aware that family circumstances can be difficult and it may not be possible to obtain parental samples in some cases. Interpretation of an imbalance detected on array CGH is less informative without inheritance studies. Will array CGH detect mutations? No. The array platform we will be using will detect small genomic imbalances which may disrupt genes and give a similar phenotype to a mutation but it is not sensitive enough to detect mutations within genes. Requests for specific mutation testing can be made to the Leeds DNA laboratory. Which referral card do I use? It's the Cytogenetics and DNA Joint referral card which you already use. It is important that any patient referred for array CGH testing is appropriately consented and counselled about the technology, its limitations, target turnaround times, and reporting arrangements. Document1