Water Structure and Acid-Base Equilibrium
advertisement

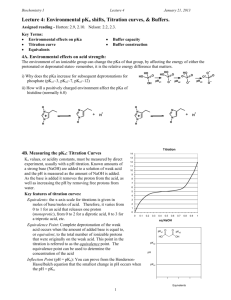
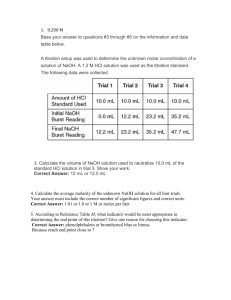
Biochemistry I Lecture 4 August 30, 2015 Lecture 4: Environmental pKa shifts, Titration curves, & Buffers. Goals: Predict effect of environment on pKa Obtain pKa from a titration curve. Draw a titration curve Understand the molecular nature of buffers. Select correct weak acid for a buffer Buffer construction 4A. Environmental effects on acid strength: The environment of an ionizable group can change the pKa of that group, by affecting the energy of either the protonated or depronated states- remember, it is the relative energy difference between the HA and A- states that matters. If the HA state is stabilized relative to A-, the acid is weaker, it prefers to stay protonated. If the HA state is destabilized, the acid will be stronger, since it prefers to be deprotonated. If the A- state is stabilized, the acid will be stronger since it prefers to deprotonate If the A- state is destabilized, the acid will be weaker since it prefers to remain protonated. How will a positively charged environment affect the pKa of histidine (normally its pKa=6.0) E>0 E=0 E<0 HA A HA A A HA Weaker Stronger 4B. Measuring the pKa: Titration Curves Ka values, or acidity constants, must be measured by direct experiment, usually with a pH titration. Known amounts of a strong base (NaOH) are added to a solution of weak acid and the pH is measured as the amount of NaOH is added. As the base is added it removes the proton from the acid, as well as increasing the pH by removing free protons from water. Key features of titration curves: Equivalents: the x-axis scale for titrations is given in moles of strong base/moles of weak acid. Therefore, it varies from 0 to 1 for an acid that releases one proton (monoprotic), from 0 to 2 for a diprotic acid, 0 to 3 for a triprotic acid, etc. Equivalents of NaOH (bottom scale) when going from left to right. This is also the same as fA- since you are starting with the fully protonated acid (fa-=0). Equivalents of HCl when going from right to left (top scale on graph). This is also the same as fHA since you are starting with fully deprotonated acid (fHA = 0) 1 Biochemistry I Lecture 4 August 30, 2015 Equivalence Point (Eq Pt): Complete deprotonation of the weak acid occurs when the amount of added base is equal to, or equivalent, to the total number of ionizable protons that were originally on the weak acid. This point in the titration is referred to as the equivalence point. The equivalence point can be used to determine the concentration of the acid Inflection Point (pH = pKa): You can prove from the Henderson-Hasselbalch equation that the smallest change in pH occurs when the pH = pKa. 4C. Buffers: A pH buffer is an acid that resists changes in the solution pH by absorbing or releasing protons. Buffers play an important role in cellular processes because they maintain the pH at an optimal level for biological processes. They are also widely used to control pH in laboratory processes. pH Titration 14 14 13 13 12 12 11 11 10 10 9 9 8 8 7 7 6 6 5 5 4 4 3 3 2 2 1 1 0 0 0 0.2 0.4 0.6 0.8 1 0 0.1 Fraction Protonated 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 eq NaOH Buffering range/region: Buffering capacity: Total moles of a strong acid or base that can be absorbed by a buffer solution and keep the pH within the buffer region. The maximum buffer capacity is, approximately: 0.6 x [AT] This corresponds to starting at one end of the buffer region and adding base (left to right) or acid (right to left) to go to the other end of the buffer region. Making a Mono-protic buffer with concentration [AT] and volume V. [AT] = [HA] + [A] Need to determine the ratio of [A-] to [HA] to give desired pH. This can be calculated from the Henderson-Hasselbalch equation since the pH is defined, and so is the pKa of the acid. 2 [ A ] pH pK A log [ HA] 1 Biochemistry I Lecture 4 August 30, 2015 pH Method: 1. Select a weak acid whose pKa is within one pH titration pH unit of the desired pH. 9 2. Determine the fraction protonated and deprotonated at the desired pH, fHA & fA-. 8 3. Obtain this ratio of [HA] to [A-] by one of 7 the following three methods: 1 i) Mix the indicated concentration of the 6 weak acid and its conjugate base (e.g. 5 sodium salt) to give the desired pH: 4 moles (HA)= fHA [AT]× V moles (A-) = fA3 [AT] × V 2 ii) Use [AT] amount of the acid form of the weak acid and add sufficient strong base 1 (e.g. NaOH) to make the required 0 concentration of [A-] to attain the desired 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 pH. You are titrating starting from the left Equivalents side and converting enough of the fully protonated acid to give the correct amount of the deprotonated acid. The added base converts HA to A-. The amount of strong base to add is fA- equivalents. moles NaOH = fA- × [AT] × V This would be added to AT×V moles of the weak acid. 1 iii) Use [AT] amount of the conjugate base form of the weak acid and add sufficient strong acid (e.g. HCl) to make the required concentration of [HA] to attain the desired pH. You are protonated the fully deprotonated acid by just the right amount to give the correct amount of the protonated acid. The added acid converts A- to HA. The amount of strong acid to add is fHA equivalents. moles HCl = fHA × [AT] × V This would be added to AT×V moles of the weak acid.. Example: Make 1L of 1 M buffer solution at pH 5.0 using one of the following two buffers. Buffer Histidine Pyruvic Acid pKa pH titration 9 6.0 (sidechain) 2.50 8 7 R 10 ( pH pKa ) 1 f HA 1 R 6 pH 1. Which buffer would you use and why? 2. Determine fraction protonated and deprotonated at the desired pH 5 4 3 f A R 1 R 2 1 0 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1 pHEquivalents titration 9 3. Obtain the desired ratio of [HA] and [A-] i) Mixing the appropriate amount of the acid and base form of the buffering acid. 8 7 pH 6 5 4 ii) Starting with AT x V moles of the pure weak acid, HA, add fAequivalents of a strong base, or fA- [AT] × V moles of the strong base. 3 2 1 0 0 1 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 Equivalents In practice, the number of moles of NaOH or HCl is not directly measured, rather a pH electrode is used to monitor the pH of the solution, and sufficient NaOH or HCl is added until the desired pH is reached. 3 1 Biochemistry I Lecture 4 August 30, 2015 pH titration iii) Starting with AT x V moles the pure sodium salt, NaA, add fHA equivalents of a strong acid, or fAH [AT] × V moles of the strong acid. 9 8 7 pH 6 5 4 3 2 1 0 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1 Equivalents Polyprotic Buffers: a) Use the pKa that is closest to the desired pH. b) Calculate fHA and fA- using that pKa. c) Generate buffer by: i) Mix the required molar amounts of “HA” and “A” forms of the buffer, where HA is the acid form for the pKa that you used. AH2 AH A ii) Start with the fully protonated weak acid and add x equivalents of NaOH, where x = n + fA-, and “n” is the number of buffer regions you have to cross, beginning at left of titration curve, to get to the region you used. iii) Start with the fully deprotonated acid and add x equivalents of HCl, where x = n + fHA , and “n” is the number of buffer regions you have to cross, beginning at the right of the titration curve, to get to the region you used. Diprotic buffer example: The diprotic acid has pKa values of 3 and 6. How would [ A ] 1 R you make 0.5 L of a 0.25 M buffer solution at pH=2.5? R 10( pH pKa) f HA fA- [ HA] 1 R 1 R Step a: Pick the ionization whose pKa is closest to the desired pH 1 0.33 ( 2.5 3.0) Step b: Use that pKa to calculate the fraction of each R 10 1.33 1.33 species, fHA and fA 0.33 0.75 0.25 Step c: Method i: moles AH = fA- × [AT] x V = 0.25 × 0.25 moles/L × 0.5 L = 0.03125 moles moles AH2 = fHA × [AT] x V = 0.75 × 0.25 moles/L × 0.5 L = 0.09375 moles Method iii – using the disodium salt. eq. of HCl = 1 + 0.75 = 1.75 eq moles of HCl = 1.75 [AT]V = 1.75 × 0.25 moles/L × 0.5 L = 0.218 moles HCl. This would be added to [AT] × V moles of Na2A = 0.125 moles. pH 10 9 8 7 6 5 4 3 2 1 0 0 0.5 1 Equivalents 4 1.5 2