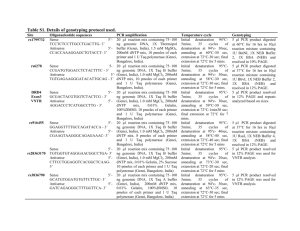
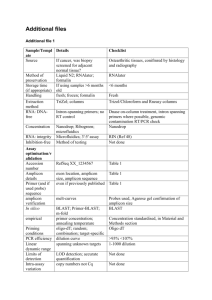
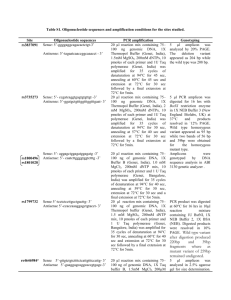
Table S1. Protocol followed for amplification of target sites. Gene
advertisement

Table S1. Protocol followed for amplification of target sites. Gene Site rs2236225 MTHFD (C/T) rs1051266 RFC1 (G/A) rs1801131 (A/C) Amplicon Primer sequence 5′-3′ Reaction mixture for PCR Temperature cycle Sense: TCCCAATTCCC CTGATGTTA. Antisense: AGGGCACTCCA GTGTTTGTC. Total volume 20 μl: 75100ng template DNA, 20 pmoles of each primer, 1.0 U Taq polymerase, 200 µM dNTP mix, 1X Thermopol buffer (Genei) and 2.5 mM MgSO4. Initial denaturation 95° - 5 min; 35 cycles of denaturation: 95°C - 35 s, annealing: 60°C - 35 s and extension: 72°C - 35 s; final extension 72°C - 5 min 179 Sense: AGGCACAGTGT CACCTTCGT. Antisense: TGAAGCCGTAG AAGCAAAGG. Total volume 20 μl: 75100ng template DNA, 20 pmoles of each primer, 1.0 U Taq polymerase, 200 µM dNTP mix, 1X buffer B(Genei) and 1.0 mM MgCl2. Initial denaturation 95° - 5 min; 35 cycles of denaturation: 95°C - 30 s, annealing: 59°C - 30 s and extension: 72°C - 30 s; final extension 72°C - 5 min 163 Sense: TTTGGGGAGCT GAAGGACTA. Antisense: CTTTGTGACCA TTCCGGTTT. 20 μl mix containing 75100ng template DNA, 20 pmoles of each primer, 1.0 U Taq polymerase, 200 µM dNTP mix, 1X buffer B (Genei) and 1.5 mM MgCl2. Initial denaturation 95° - 5 min; 35 cycles of denaturation: 95°C - 35 s, annealing: 60°C - 35 s and extension: 72°C - 35 s; final extension 72°C - 5 min 160 Sense: GAAGCAGGGA GCTTTGAGG. Antisense: ACGATGGGGCA AGTGATG-3′. Total volume 20 μl: 75100ng template DNA, 20 pmoles of each primer, 1.0 U Taq polymerase, 200 µM dNTP mix, 1X buffer B (Genei) and 1.0 mM MgCl2. Initial denaturation 95° - 5 min; 35 cycles of denaturation: 94°C - 30 s, annealing: 56°C - 30 s and extension: 72°C - 30 s; final extension 72°C - 5 min 152 size (bp) MTHFR rs1801133 (C/T) Table S2. Detailed description for the Restriction Fragment Length Polymorphism analysis. ID rs2236225 (C/T) rs1051266 (G/A) rs1801131 (A/C) rs1801133 (C/T) Analytic details Final volume 20l. 5 μl PCR amplicon was digested with 1U of MspI and 1X NEBuffer 4 at 37C for overnight. Products resolved by 12% PAGE. Final volume 20l. 3 μl PCR amplicon was digested with 1U of SfoI and 1X NEBuffer 4 at 37C for overnight. Products resolved by 12% PAGE. Final volume 20l. 5 μl PCR amplicon was digested with 1U MboII (NEB, USA) and 1X NEB buffer 4 at 37C for 3hrs. Products were resolved in 20% PAGE. Final volume 20l. 5 μl PCR amplicon was digested with 1U of HinfI (NEB, USA) and 1x New England Biolabs Buffer 4 at 37C for 3hrs. Products were resolved in 12% PAGE. Recognition sites Genotyping 5'-C▼CGG-3' Wild-type CC, heterozygous CT and homozygous mutant TT. Two fragments of 103 and 76bp from a 179bp product that carried a C>T mutation. 3'-GGC▲C-5' 5'-GGC▼GCC-3' 3'-CCG▲CGG-5' 5'-GAAGA(N)8▼-3' 3'-CTTCT(N)7▲-5' 5'-G▼ANTC-3' 3'-CTNA▲G-5' Wild-type GG, heterozygous GA and homozygous mutant AA. Two fragments of 130 and 33bp from a 163bp product that carried a G>A mutation. Wild-type AA, heterozygous AC and homozygous mutant CC. AA produced four fragments of 54, 30, 28 and 18bp, CC genotype yielded three fragments of 82, 30 and 18bp from 160bp product due to abolition of one restriction site. Wild-type CC, heterozygous CT and homozygous mutant TT. Two fragments of 96 and 56bp from a 152bp product that carried a C>T mutation. Table S3. Comparative analysis of genotype frequencies in female probands and control . Gene Site ID MTHFD rs2236225 (C/T) RFC1 rs1051266 (G/A) rs1801131 (A/C) MTHFR rs1801133 (C/T) Genotype Control (N=20) Proband (N=18) CC CT TT GG GA AA AA 0.18 0.51 0.31 0.25 0.53 0.22 0.29 0.28 0.33 0.39 0.17 0.39 0.44 0.22 AC 0.53 0.67 CC 0.18 0.11 CC 0.74 0.56 CT 0.23 0.44 TT 0.03 0.00 2 (p-value) 6.95 (0.031) 11.0 (0.004) 4.28 (0.117) 12.1 (0.002) Table S4. Comparative analysis of haplotype frequencies using case-control data. Control Gene Sites Case Groups Haplotype 2 (p-value) OR A-C 0.42 0.44 0.1909 (0.6622) 1.0 A-T 0.44 0.44 0.0006698 (0.9794) 0.98 C-C 0.12 0.11 0.1599 (0.6892) 0.92 C-T 0.02 0.01 1.068 (0.3013) 0.46 A-C 0.42 0.45 0.2331 (0.6292) 1.0 A-T 0.44 0.44 0.0009066 (0.976) 0.94 C-C 0.13 0.10 0.4685 (0.4937) 0.79 C-T 0.01 0.01 0.01903 (0.8903) 0.95 A-C 0.42 0.34 0.249 (0.6177) 1.0 A-T 0.43 0.44 0.1069 (0.7437) 1.26 C-C 0.12 0.21 1.219 (0.2695 ) 2.334 C-T 0.03 1.064e009 0.2466 (0.6195) 4.282e-008 All rs1801131 MTHFR - Male rs1801133 Female Table S5. Analysis of gene-gene interaction by Multi Dimensionality Reduction using case-control dataset. Group All probands & controls Male subjects Female subjects Model TBA Prediction Error CVC P- value Power 1 0.4929 0.5071 7/10 0.8880 100% 23 0.5222 0.4778 7/10 0.6440 100% 124 0.5369 0.4631 9/10 0.4710 100% 1234 0.4701 0.5299 10/10 0.9810-0.9820 100% 1 0.5727 0.4273 10/10 0.2070-0.2080 100% 14 0.5439 0.4561 6/10 0.4720-0.4730 100% 124 0.6102 0.3898 10/10 0.0360-0.0370 100% 1234 0.5372 0.4628 10/10 0.5330 100% 3 0.6000 0.4000 8/10 0.5570 100% 23 0.6333 0.3667 6/10 0.4640 100% 234 0.5278 0.4722 7/10 0.7690 100% 1234 0.6944 0.3056 10/10 0.2500-0.2510 100% NB. 1- rs2236225, 2- rs1051266, 3-rs1801131, 4-rs1801133; Testing Balance Accuracy- TBA; Cross validation consistency-CVC.