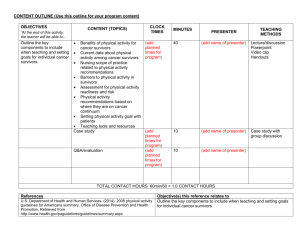
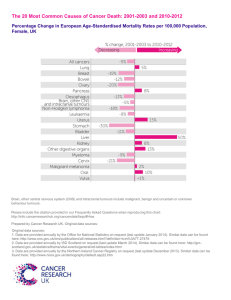
GBM-protein-lay-summary - Brain Tumour Research Campaign
advertisement

Glioblastoma multiformae is an aggressive form of astrocytoma (cancer of the astrocyte cells that biochemically support to the brain’s neurons). It accounts for about 50% of all brain tumours, with an average patient survival time of around one year even with surgical intervention, radiotherapy and chemotherapy. Despite this poor prognosis, 36% of patients remain alive after 3 years and are called “long-term survivors”. It might be expected that this small cohort of long-term survivors would be readily identifiable in terms of the clinical characteristics of their tumour. However, despite a number of research efforts, no distinct markers have been found. Pathologists are therefore unable to reliably predict which patients will be long-term survivors and the mechanisms behind their long-term survival remain unknown. If the molecular differences between the tumours from sort-term and long-term survivors could be established, treatment plans could be better tailored towards individual patients and new drugs to prolong life could potentially be developed. In molecular biology, genes encoded in an individual’s genetic blueprint (DNA) are converted into their functional products (proteins) via an intermediate molecule called RNA. To date, research aimed at determining gene expression abnormalities has focussed on analysing DNA or RNA. This project is different in that it focuses at the level of the functional cellular proteins rather than at the genetic level, asking the question “what is different in terms of protein expression in tumours obtained from short-term survivors and long-term survivors of glioblastoma multiforme?” The protein content of tumour biosamples from short-term and long-term survivors will be analysed using a technique called “two-dimensional difference gel electrophoresis”. This involves resolving the protein complement of the tumours into thousands of distinct spots on A4-sized gels, each spot containing a different protein. The spot patterns from each biosample are compared and the spots containing differentiallyexpressed proteins highlighted. Although this description of how different the tumours from short- and long-term survivors are is valuable, the identities of the proteins still need to be established. This is done by cutting the differentially-expressed spots from the gels, extracting the protein from them and analysing it by mass spectrometry. Mass spectrometry determines the amino acid sequence of each protein and this is used to establish its identity. By understanding the key biological differences at the protein level between tumours that kill the patient quickly, and those that do not, we aim to identify key proteins that could be targeted by either existing or novel drugs, to improve the prognosis for patients that have a biologically aggressive tumour. Although we accept that this may not result necessarily in a cure for an individual patient, it has the potential to increase life expectancy for the group of patients that have the worst prognosis.