Supplementary Figures - Springer Static Content Server
advertisement
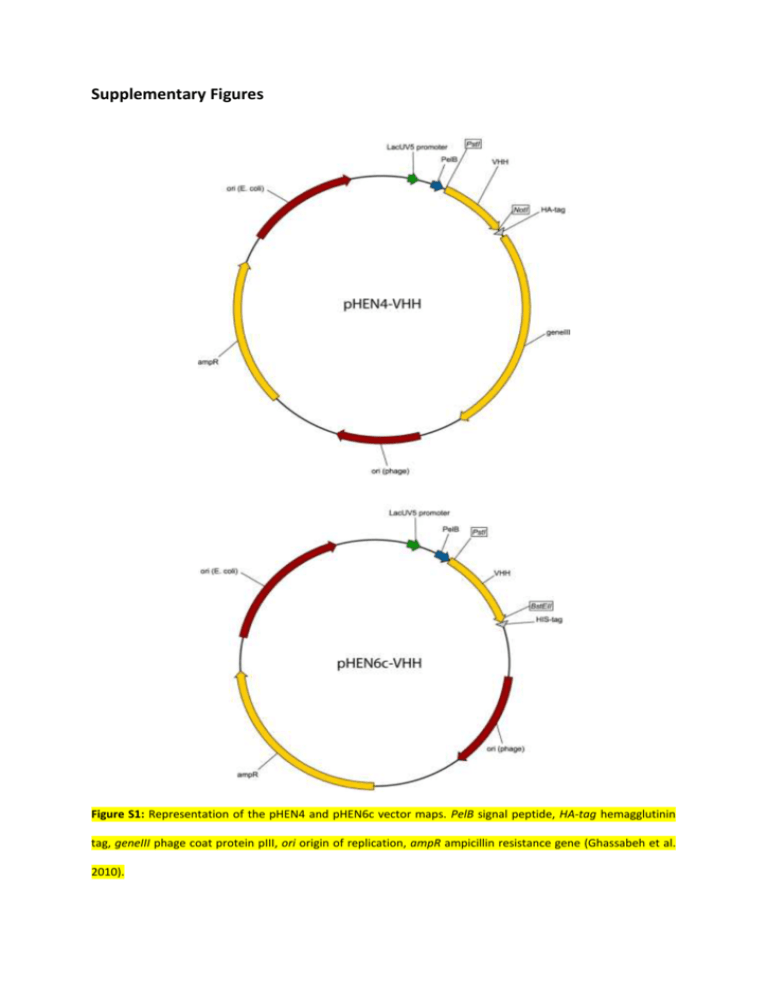
Supplementary Figures Figure S1: Representation of the pHEN4 and pHEN6c vector maps. PelB signal peptide, HA-tag hemagglutinin tag, geneIII phage coat protein pIII, ori origin of replication, ampR ampicillin resistance gene (Ghassabeh et al. 2010). Figure S2: The in E. coli produced and IMAC purified VHHs were analysed on SDS-PAGE (15%). Figure S3: The VHHs were analysed as primary antibodies in western blot. After blotting 34 µg of Arabidopsis seed protein extract under reducing conditions, mixtures of VHHs were tested as primary antibodies, followed by mouse anti-his and sheep anti-mouse antibodies fused with horse radish peroxidase. In each western blot (only three are depicted here) one aspecific band (arrowheads) was visualised. As a control for the secondary and tertiary antibodies, 30 ng of VHH30 was loaded (arrows). Figure S4: Alignment of albumin protein sequences of the Brassicaceae family. Several albumin protein sequences were aligned using ClustalW. All albumin protein precursors contain an N-terminal signal peptide, an N-terminal processed peptide, a small albumin subunit (yellow), an internal linker region, a large albumin subunit (blue) and a C-terminal processed peptide. The subunit assignments for At_SESA1, 2, 3 and 4 and Bn_napA were based on Krebbers et al. (1988). The subunit boundaries of the other albumin homologues have been deduced via protein alignment. However, for At_SESA5 no clear assignment was possible. The reference papers of all the sequences are listed: Sa Sinapis alba (Delapena et al. 1993), Bn Brassica napus (Baszczynski and Fallis 1990; Ericson et al. 1991; Josefsson et al. 1987), Rs Raphanus sativus (Raynal et al. 1991), At Arabidopsis thaliana (Krebbers et al. 1988; van der Klei et al. 1993). Supplementary References Baszczynski CL, Fallis L (1990) Isolation and nucleotide sequence of a genomic clone encoding a new Brassica napus napin gene. Plant Mol Biol 14 (4):633-635. doi:10.1007/BF00027511 Delapena MAG, Villalba M, Garcialopez JL, Rodriguez R (1993) Cloning and expression of the major allergen from yellow mustard seeds, Sin a I. Biochem Biophys Res Commun 190 (2):648-653. doi:10.1006/bbrc.1993.1097 Ericson ML, Murén E, Gustavsson H-O, Josefsson L-G, Rask L (1991) Analysis of the promoter region of napin genes from Brassica napus demonstrates binding of nuclear protein in vitro to a conserved sequence motif. Eur J Biochem 197 (3):741-746. doi:10.1111/j.14321033.1991.tb15966.x Josefsson L-G, Lenman M, Ericson ML, Rask L (1987) Structure of a gene encoding the 1.7 S storage protein, napin, from Brassica napus. J Biol Chem 262 (25):12196-12201 Krebbers E, Herdies L, De Clercq A, Seurinck J, Leemans J, Van Damme J, Segura M, Gheysen G, Van Montagu M, Vandekerckhove J (1988) Determination of the processing sites of an Arabidopsis 2S albumin and characterization of the complete gene family. Plant Physiol 87 (4):859-866. doi:10.1104/pp.87.4.859 Raynal M, Depigny D, Grellet F, Delseny M (1991) Characterization and evolution of napin-encoding genes in radish and related crucifers. Gene 99 (1):77-86. doi:10.1016/0378-1119(91)90036-B van der Klei H, Van Damme J, Casteels P, Krebbers E (1993) A fifth 2S albumin isoform is present in Arabidopsis thaliana. Plant Physiol 101 (4):1415-1416. doi:10.1104/pp.101.4.1415 Supplementary Table legend Table S1: The VHH antigen identification details were obtained with the 4800 MALDI TOF/TOF Proteomics Analyzer (AB SCIEX) and the GPS explorer v3.6 (AB SCIEX) software package combined with search engine Mascot version 2.2 (Matrix Science). Protein score, The score calculated by the Mascot search engine for each protein. This score is based on the probability that peptide mass matches are non-random events. If the protein score is equal to or greater than the Mascot® Significance Level calculated for the database search, the protein match is considered to be statistically non-random at the 95% confidence interval. Protein score = - 10 * Log (P), where P is the probability that the observed match is a random event. Expect, Protein score expectation value. RMS error (ppm), RMS error of the set of matched mass values, in ppm. Sequence coverage %, Percentage of protein sequence covered by assigned peptide matches. Unique peptides, The number of peptides with unique sequences matching the selected protein. Total Ion Score, A score calculated by weighting ion scores for all individual peptides matched to a given protein. Peptide Number, Peptide index number within the list of peptides associated with a given protein. Observed, The observed monoisotopic mass of the peptide in the spectrum (m/z). Mr (Exp), The experimental mass of the peptide calculated from the observed m/z value. Mr (Calc), The theoretical mass of the peptide based on its sequence. Delta (Da), The difference between the theoretical (Mr (Calc)) and experimental (Mr (Exp)) masses, in dalton. Start, The starting position of the peptide in the protein. End, The ending position of the peptide in the protein. Miss, The number of missed trypsin cleavage sites. Ions score, The ions score is calculated by the Mascot search engine for each peptide matched from MS/MS peak lists. This score is based on the probability that ion fragmentation matches are non-random events. If the ion score is equal to or greater than the Mascot® Significance Level calculated for the database search, the peptide match is considered to be statistically non-random at the 95% confidence interval. Ions score = - 10 * Log (P), where P is the probability that the observed match is a random event. Best Ions score, The highest individual Ion score for a given protein identification. Expect, Ions score expectation value. Peptide, The amino acid sequence of the selected peptide. Variable Modification Variable modification type on the peptide.









