Supplementary Information (docx 40K)
advertisement

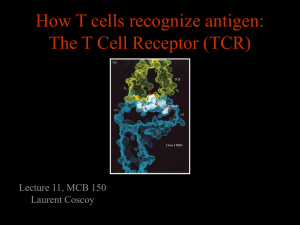
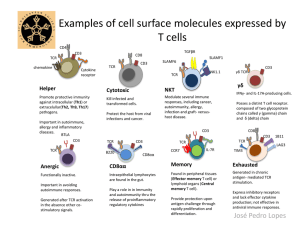
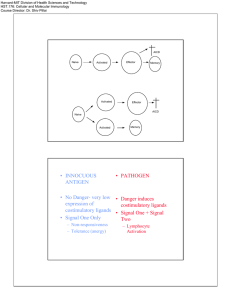
Supplemental Materials Metal-specific CD4+ T cell responses induced by beryllium exposure in HLA-DP2 transgenic mice Michael T. Falta, PhD1, Alex N. Tinega1, Douglas G. Mack, PhD1, Natalie A. Bowerman, PhD1, Frances Crawford2,3, John W. Kappler, PhD2,3, Clemencia Pinilla, PhD4, and Andrew P. Fontenot. MD1,2 Departments of 1Medicine and 2Immunology, University of Colorado Anschutz Medical Campus, Aurora, CO 80045 USA 3 Howard Hughes Medical Institute, National Jewish Health, Denver, CO 80206 USA 4 Torrey Pines Institute for Molecular Studies, San Diego, CA 92121 USA 1 Supplemental Methods Flow cytometric analysis of TCR Vβ expression Prepared splenocytes, lung and BAL cells isolated from unexposed or BeO-exposed HLA-DP2 Tg FVB/N mice were stained either individually or as pooled samples with fluorescently conjugated mAb specific to CD4 (PerCP or APC) and murine TCR Vβ (FITC). The panel of anti-TCR Vβ mAb (BD Pharmingen) included anti-mouse Vβ2 (clone B20.6), Vβ3 (KJ25), Vβ4 (KT4), Vβ6 (RR4-7), Vβ7 (TR310), Vβ14 (14-2), and Vβ17 (KJ23). Cells were run on a FACSCalibur flow cytometer (BD), and the frequency of CD4+ T cells in the lymphocyte scatter gate that expressed the different Vβ’s was assessed using FlowJo software (Tree Star). Transfectomas expressing particular murine Be-specific TCRs TCRs expressed by select Be-specific hybridomas were cloned into expression vectors and stably transfected into the mouse hybridoma cell line 54ζ, which expresses human CD4 (1-4). Separate expression vectors encoding the variable domains of the TCRA and TCRB genes juxtaposed with either the murine AC or BC gene segments were generated and co-transfected into recipient cells by electroporation. Transfectants were selected in G418-containing medium (1 mg/ml; Cellgro), screened for TCR expression using a PE-labeled H57-597 mAb (eBioscience), and cloned by limiting dilution to identify subclones with the highest TCR expression (data not shown). HLA-DP2 transfected fibroblast cell lines Murine fibroblasts transfected with genes encoding wild-type HLA-DP2 or HLA-DP2 with single amino acid substitutions in the β-chain were used as antigen-presenting cells to present Be and peptides. DP2.21 cells are kidney fibroblasts derived from IAb-/-, Ii-/- C57BL/6 mice (5) and co- 2 transfected to express HLA-DP2 (DPA*0103/DPB1*0201). For T cell transfectoma activation assays, this cell line was adapted to grow in protein-free tissue culture medium (OptiPro SFM; Invitrogen), supplemented with 4 mM glutaMAX (Invitrogen). DP8302 is a DAP.3 L cell line that also expresses wild-type HLA-DP2 (6, 7). For HLA-DP2 β-chain variants, site-directed mutagenesis of the DPB1*0201 gene using overlapping PCR primers to encode single amino acid gene mutations was performed as previously described (6, 8). HLA-DP2 variants were expressed on the DAP.3 L cell line and stained with an anti-HLA-DP mAb (clone B7.21) to ensure similar surface expression as wild-type (data not shown). All DAP.3 L cell lines were grown in complete culture media (C-IMDM) consisting of IMDM/glutamax tissue culture medium (Invitrogen) supplemented with 10% heat-inactivated FBS (Hyclone), 1 mM sodium pyruvate, 100 units/ml penicillin, 100 µg/ml streptomycin (all from Invitrogen). T cell hybridoma/transfectoma activation assays T cell hybridomas or transfectomas (1 x 105 cells per well) were cultured in 96-well flat-bottomed tissue culture plates (Falcon) in the presence of HLA-DP2-transfected murine fibroblasts (1 x 105 cells) and varying amounts of BeSO4 and peptides. For screening of the decapeptide positional scanning library, mixtures were added (200 µg/ml) in duplicate to DP2.21 cells in protein-free medium for 3 hours prior to the addition of T cell transfectoma cells and BeSO4 (75 µM). Identical experimental conditions were used to test crude preparations of mimotopes (0.5, 5.0 and 50 g/ml) as well as for performing dose response curves to purified peptides (0.1 nM to 10 M) in the presence of BeSO4. Standard dose response curves to BeSO4 (range concentrations) and analysis of transfectoma response to HLA-DP2 variants presenting different pure peptides were performed in C-IMDM. 3 Cloning of PLXNA4/Be-specific TCRs from CD4+ T cells isolated by single cell sorting A single cell suspension was prepared from the lungs of BeO-exposed HLA-DP2 Tg mice as described previously (9). Cells were stained with mAbs directed against CD4 (AF700), CD3 (PECy7), CD8 (eFluor 450), B220 (eFluor 450), F4/80 (eFluor 450), CD44 (PerCP-Cy5.5), and PElabeled HLA-DP2-PLXNA4/Be tetramer (4). Tetramer-staining CD3+CD4+CD44+ T cells were single cell sorted into reverse transcription buffer in a 96-well plate using a FACSAria flow cytometer. TCRAV and TCRBV gene expression was determined using 5’ RACE and a nested PCR method as previously described (3, 10). In some experiments, tetramer-staining T cells were sorted directly into complete IMDM media in a 96 well round-bottomed plate and stimulated with anti-CD3 and anti-CD28 beads (Invitrogen) and recombinant human IL-2 (30 U/mL) for two weeks. Stimulated cells were transferred into a 24 well plate for further stimulation and expansion. The TCRAV and BV gene expression in the expanded cells was determined as described above. T cell transfectomas expressing HLA-DP2-PLXNA4/Be binding TCRs were generated as described previously (2, 4). Cells expressing the highest levels of TCR as detected by staining with anti-mouse TCR β-PE (H57-597) mAb (BD Biosciences) were purified by cell sorting and screened for Be specificity using an IL-2 ELISA. In all experiments, supernatants were harvested after 22-24 hrs, and murine IL-2 was measured by ELISA (eBioscience). For dose response calculations, the concentration of peptide that provided 50% of the maximum IL-2 response (EC50) for each hybridoma was determined using nonlinear regression (sigmoidal-fit; Prism, GraphPad Software, Inc.). 4 Supplemental Figure Legends Figure S1. TCRA gene conferring Be-specificity for T cell hybridomas LB9-18 and LB10-17. Three Be-specific T cell hybridomas derived from the lungs of BeO-exposed HLA-DP2 Tg mice expressed multiple TCR α-chains paired with a TCR Vβ6-chain (LB9-18/LB10-9 are clonal replicates and LB10-17; see Figure 1B). To ascertain which TCR α/β pair conferred Be-specificity for each hybridoma, the TCRA genes from each hybridoma were introduced separately into transfectomas expressing the appropriate TCRB gene. TCR surface expression on the resultant transfectomas was confirmed by staining with an anti-mouse TCR β-chain mAb (H57-597) (data not shown). Transfectomas were tested for their ability to secrete IL-2 (mean ± SEM pg/ml) in response to BeSO4 (200 µM) presented by HLA-DP2 transfected fibroblasts. Shown are the correct TCR α/β pairs for each hybridoma (Vβ6/Vα4.2 for LB9-18; Vβ6/Vα1.1 for LB10-17) as indicated by IL-2 production. Figure S2. T cell transfectoma response to mimotope 6 presented by transfected DAP3.L cell lines expressing wild-type or variant HLA-DP2 genes. LB9-18 and LBM10-10 cells (1 x 105 cells per well) were mixed in complete IMDM with an equal number of DAP3.L fibroblast cells transfected with either wild-type (8302), or variant HLA-DP2 molecules containing single-site mutations at critical Be-coordinating residues (amino acid positions 26, 68 and 69) of the HLADP2 β-chain. Cells were stimulated with either Be (200 µM) or mimotope 6 (5 µM) for 22 hours and IL-2 secretion (mean ± SEM pg/ml) measured by ELISA. Data shown are representative of three independent experiments done in triplicate. 5 References 1. Boen E., Crownover A.R., McIlhaney M., Korman A.J. & Bill J. Identification of T cell ligands in a library of peptides covalently attached to HLA-DR4. J Immunol 165, 2040-2047 (2000). 2. Bowerman N.A., Falta M.T., Mack D.G., Kappler J.W. & Fontenot A.P. Mutagenesis of beryllium-specific TCRs suggests an unusual binding topology for antigen recognition. J Immunol 187, 3694-3703 (2011). 3. Bowerman N.A., et al. Identification of multiple public T cell receptor repertoires in chronic beryllium disease. J Immunol 192, 4571-4580 (2014). 4. Falta M.T., et al. Identification of beryllium-dependent peptides recognized by CD4+ T cells in chronic beryllium disease. J Exp Med 210, 1403-1418 (2013). 5. Huseby E.S., Crawford F., White J., Kappler J. & Marrack P. Negative selection imparts peptide specificity to the mature T cell repertoire. Proc Natl Acad Sci U S A 100, 11565-11570 (2003). 6. Bill J.R., et al. Beryllium presentation to CD4+ T cells is dependent on a single amino acid residue of the MHC class II b-chain. J Immunol 175, 7029-7037 (2005). 7. Fontenot A.P., Torres M., Marshall W.H., Newman L.S. & Kotzin B.L. Beryllium presentation to CD4+ T cells underlies disease susceptibility HLA-DP alleles in chronic beryllium disease. Proc Natl Acad Sci U S A 97, 12717-12722 (2000). 8. Dai S., et al. Crystal structure of HLA-DP2: Implications for chronic beryllium disease. Proc Natl Acad Sci U S A 107, 7425-7430 (2010). 6 9. Mack D.G., et al. Regulatory T cells modulate granulomatous inflammation in an HLA- DP2 transgenic murine model of beryllium-induced disease. Proc Natl Acad Sci U S A 111, 85538558 (2014). 10. Ozawa T., Tajiri K., Kishi H. & Muraguchi A. Comprehensive analysis of the functional TCR repertoire at the single-cell level. Biochem Biophys Res Commun 367, 820-825 (2008). 7