Atomic Orbital Overlap
advertisement

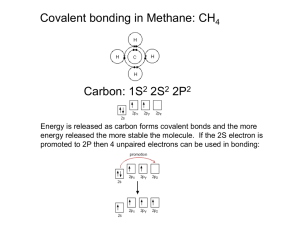
CBB NOTES – Chemistry and Biochemistry of Molecules Lecture 1 – Atoms and Molecules 1 Recall the fundamental elements of atomic structure, their properties and organisation Protons positively charged. Neutrons no charge Protons and neutrons have approx. the same mass. Nucleus: positively charges, contains protons and neutrons. ATOMIC NUMBER (Z) – number of protons in the nucleus. Defines the element. Number of protons dictates the number of electrons and therefore the chemistry of the element. ATOMIC MASS NUMBER (A) – the combined number of protons and neutrons. Neutron number = atomic mass number – atomic number Electrons negatively charged, mass approx. 1/2000 of a proton. Number of electron in nucleus = number of protons in nucleus. AMU (Atomic Mass Unit): 1/12 of the mass of an atom of carbon-12. Aka RMM or Dalton Isotopes: Isotopes of an element have the same atomic number (Z – number of protons) but differ in number of neutrons so have different atomic mass numbers (A – protons and neutrons) As they have the same atomic number, they have same number of electrons and so similar chemistry. Atomic masses are measured/calculated relative to carbon-12 MOLES: 1 moles of a substance has the same number of things as there are atoms in 12g of carbon12 Avogadro’s number = 6.022 x 1023 mol-1 Molar mass = mass in grams of 1 mole of substance A atom of carbon-12 weight 12amu. 12g gives 1 mole of carbon 12. MOLECULAR MASS (M): the sum of the atomic masses of the atoms represented in the molecular formula. FORMULA WEIGHT (FW): more suitable when not in a molecule. Compounds crystallised with water of hydration, the water must be included in FW. MOLARITY: describes the concentration of a solution. e.g. one mole of substance in 1 litre of water is a one molar solution. 1 mol dm-3 Describe the shape, number and relative energies of s, p and d orbitals Quantum mechanics and Schrodinger The classical model assumed electrons move in trajectories around then nucleus (Rutherford-Bohr model) Electrons are instead considered to have wave-like properties. Schrodinger’s wave equation. Solutions of this are wave functions. Atomic Orbitals: give the probability of finding an electron at a given position, based on Schrodinger wave equation. Position of an electron can be visualised as a boundary surface where there’ a >90% change of finding it. Atomic orbitals are specified by QUANTUM NUMBERS: o o o o Principal (n) – specifies the shell and size of orbital. Orbitals with higher numbers extend further from nucleus. Orbital (or azimulthal) (l) – specifies subshell, determines orbital shape. Magnetic (ml) – indicated orbital orientation Spin (ms) - +1/2 or -1/2. Important when considering covalent bonds. Each orbital can hold 2 electrons with opposite spins. Within an atom, no 2 electrons can have the same set of these 4 quantum numbers. Orbitals have different shapes depending on the value of l s-orbitals SPHERICAL p-orbitals pair of drop-shaped lobes d-orbitals lobed f-orbitals multiple sets of lobes. AUFBAU PRINCIPLE – electrons occupy the lowest energy orbital first PAULI EXCLUSION PRINCIPLE – max of 2 electrons in an orbital. HUND’S RULE – electrons only pair if there’s no orbital with the same energy available. Describe ionic and covalent bonding in general terms IONIC: when a metal and a non-metal ion combine, valence electrons are transferred from the metal to the non-metal forming cations and anions that are held together by electrostatic forces COVALENT: a chemical bond formed by atoms sharing pairs of electrons. Electrons involved in covalent bonding occupy the outermost shell (valence shell). More common between non-metals Describe electronegativity and relevance to polarity of covalent bonds ELECTRONEGATIVITY: electrons not always equally shred in covalent bonds. Electronegativity measure the ability of an atom to attract the shared electrons in a bond. Greater electronegativity more strongly electrons are attracted to the atom, polarises bond. Describe hydrogen bonding and van der Waals forces VAN DER WAALS: o o Weak bonding forces in and between molecules. Intermolcular. Electrons surrounding atom/atoms in a molecule are induced to create a dipole which in turn induces further dipoles. HYDROGEN BONDING: o o o o Where hydrogen is bonded to an electronegative atom, hydrogen atom can act as a donor for a hydrogen bond with another electronegative atom. Intermolecular bond. Stronger than Van der Waals. Found in water, DNA, RNA… Give examples of the different types of bonding observed in biological systems IONIC Tertiary and quaternary structure of proteins. Ionisable R groups in amino acids, determines attraction/repulsion through spaces between residues. COVALENT Principal bond used to hold atoms in biological molecules together. E.g. amino acids, sugars, purines… SUMMARY 1. 2. 3. 4. Chemical elements are defined by their atomic number, and contain only 1 type of atom. Atoms – nucleus of protons and neutrons surrounded by electrons. Electrons are confined to orbitals of different energies and geometries. Electronic config of an atom is dictated by the number of electrons, the Aubau principle, Pauli Exclusion Principle, Hund’s rule 5. Elements can be ordered by their electrons config periodic table 6. Bonding between atoms can be covalent, ionic, hydrogen bonding, Van der Waals Lecture 2 – Atoms and Molecules 2 Describe and use the different formats of chemical notation (Lewis notation, skeletal formulae, space filling diagrams) Molecular Formula: CH4, CO2, H2O etc. Shows how the atoms are attached to each other. Structural Formula: all atoms and bonds are shown. Molecular Geometry: the 3D shape of molecules, very important to predict or understand physical properties. 3D shape can’t be predicted by Lewis theory. We can describe the shape of a molecule in terms of geometric figures which describe the arrangement of atoms around a central atom. VSEPR: VALENCE SHELL ELECTRON PAIR REPULSION A simple theory to predict molecular structures, based on assumption that valence shell electron pairs surrounding a central atom repel each other and are arranged in space in a way as to be as far away from each other as possible. e.g. Linear, tetrahedral, trigonal planar, tetrahedral, trigonal bipyramidal, octahedral. Double bonds occupy more space than single bonds Lone pair electron spread out more than bonding pairs: repulsion between lone pairs is greater than between bonding pairs. When lone pair present, angles deviate from regular geometry, take up equatorial (edge) positions rather than axial (top/bottom). 2 lone pairs will be positioned on opposite sides of an octahedron, causing square planar geometry. Understand, describe and draw resonance structures Describe orbital hybridisation VALANCE BOND THEORY (VB) focuses on the orbitals involved in the formation of covalent bonds. o Overlap – interpreted in term so of the overlap of atomic orbitals. Bonding electrons localised in the region of atomic orbital overlap. o Hybridisation – the atomic orbitals for bonding may not be ‘pure’ atomic orbitals but may have hybrid character between several possible types of orbitals. o Founded by Linus Pauling. o Is easy to visualise and use. Can describe many organic molecules and in particular molecules with multiple bonds. Fails to predict some properties: Bond length and bond energies Needs to invoke resonance to explain certain molecules Can’t predict spectroscopic properties of molecules Can’t predict paramagnetic properties of molecules Atomic Orbital Overlap: For each bond there is a condition of maximum overlap that leads to max bond strength. σ- bonds: end-to-end overlap of s, p, d and f orbitals. π-bonds: formed when p, d or f orbitals overlap sideways. Overlap less likely and bonds are weaker. In methane, s and p orbital blend together to give 4 hybrid orbitals. New orbitals are called sp3 hybrids and are equivalent to one another and point to corners of a tetrahedron. HYBRID ORBITALS: have a small lobe and a large lobe. sp2 hybrid orbitals formed too. E.g. BF3 sp hybrid orbitals…e.g. BeCl2 DOUBLE BONDS: One sigma bond formed by end on overlap of 2 py orbitals. Electron densitiy along the line connecting 2 nuclei. One formed by side on overlap of pz orbitals. Electron density above and below the line connecting the 2 nuclei Π-bond geometry Maximum overlap between pz orbitals (strongest) occurs when 6 atoms in CH2=CH2 molecule lie in the same plane. Pi bonds place strong constraints on geometry – double bonds are planar and rotation arounf the bond in energetically unfavourable. Groups connected by double bond are in a fixed position relative to each other. In aromatic molecules Pz ortbial unhybridized, half-filled orbitals available for forming pi bonds. Aromatic compounds are planar. Recall Hückel’s Rule and use it to determine whether planar ring structures will have aromatic properties HUCKEL’S RULE OF AROMACITY o It’s possible to predict whether planar rings will be aromatic o STATES that when the number of delocalised electrons (2 per double bond) is 4n + 2, where n is zero or any integer, the ring will be aromatic. Be familiar with molecular orbital diagrams and how to calculate bond order in molecules ,MOLECULAR ORBITAL THEORY Assumes the existence of molecular orbitals that are distinct from the original atomic orbitals. Molecular orbitals derive from the combination of atomic orbitals. Exist whether filled with electrons or not. E,g, H2 Molecular orbitals derive from the combination of the 2 1s atomic orbitals. Gives a σ1s (bonding orbital) and a σ*1s (antibonding orbital) Electrons fill the lowest energy orbital first. Maximum of 2 electrons with opposite spin per orbital. Advantages: 1. MO assumes there are empty orbital. Evidence provided by molecular spectroscopy techniques. 2. Implies molecular orbitals are delocalised over the molecule and doesn’t need resonance structures. 3. Can predict both bond length and bond energy. Disadvantage: can easily explain only the simpler molecules, theory very complex in larger molecules. Combination of p orbitals Describe the covalent bonds commonly found in biomolecules HOMONUCLEAR DIATOMIC MOLCULES e.g. O2. Composed of 2 atoms of the same element. HETERONUCLEAR DIATOMIC MOLCULES Molecules formed from different atoms. Electron distribution in the covalent bonds is no longer evenly shared between the 2 atoms. E.g. HF F more electronegative than H. Atomic orbitals combine to form molecular orbitals. Polar covalent bond. Charge density localised more on the more electronegative fluorine. SUMMARY In VALENCE BOND THEORY a covalent bond is interpreted in terms of the overlap of atomic orbitals. Bonding electron localised in region of orbital overlap. Atomic orbitals may not be ‘pure’. May have hybrid character in sp3, sp2, sp.. Pi and sigma bonds Double bond = 1 sigma and 2 pi bond. Triple bond = 1 sigma bond and 2 pi bonds. Planarity. Geometric constraint MOLECULAR ORBITAL THEORY assumes the existence of molecular orbitals that are derived from the combination of atomic orbitals. Combination of 2 orbitals will give bonding and anti-bonding orbitals S and p molecular orbitals Molecular orbitals are filled using same principle as filling atomic orbitals. MO theory more powerful and predicts more accurately the molecular properties but gets very complex with larger molecules. Lecture 3 – Major Classes of Biomolecules: Structure and Function Major classes Proteins - Amino Acids Carbohydrates – Saccharides and oligo-/polysaccharides Nucleotides - Nucleic Acids and DNA/RNA Lipids – Fatty Acids, Triglycerides Understand the chemical structures and properties of the major classes of biomolecules. PROTEINS AND AMINO ACIDS • Twenty Amino Acids - contain COOH and NH2 groups • α-Amino Acids as both COOH and NH2 groups attached to same carbon • α-L-Amino Acids contained in proteins/peptides • Proteins are linear chains of amino acids. • Protein chains fold in 3D due to the non-covalent interactions between regions of the linear sequence – function depends on structure CORN rule: looking down the H-Ca bond for an L amino acid we read the groups. CO-R-N clockwise ALL AMINO ACIDS IN PROTEINS ARE L STEREOISOMERS. At neutral pH amino acids exist as doubly charged Zwitterionic forms - dipolar. COO- and NH3+ Very polar molecules, high melting points Nature of side chain and pKa determines properties pKa = pH at which half ionised (CBB L4) Amino acids with uncharged side chains – free thiol (-SH) group in cysteine can form disulphide bonds. With charged side chains – can form strong H-bonds and ‘salt bridges’. Acidic contain COOK and basic contain extra NH2 groups. Carboxylate group of Glu and guanidinuim group of Arga re stabilised by resonance (way of describing delocalised electrons within certain molecules where bonding cannot be expressed by one single lewis formula). CARBOHYDRATES AND SUGARS most abundant group of biomolecules – serve as energy stores, fuels and metabolic intermediates. Many can be written as Cn(H2O)n eg Glucose is C6(H2O)6 Carbon compounds with multiple hydroxyl groups. Carbohydrates are formed by the linkage of monosaccharides (the smallest carbohydrate building blocks). 3 classes of carbohydrates: monosaccharides – simple sugars such as glucose. oligosaccharides – 2-10 monosaccharide units linked together polysaccharides – > 10 units - cellulose, glycogen, starch. MONOSACCHARIDES Can adopt a linear chain (acyclic) or a cyclic form Nucleophile addition reaction between hydroxyl group and carbonyl carbon forms a hemiacetal group. Stereocentre produced at hemiacetal group, give rise to a new stereocentre. 2 possibilities are epimers, alpha or beta depending on relative configuration. Called anomers, differences in cyclic saccharides. In solution, monoaccharides form cyclic ring structures. EPIMERS: pairs of sugars that differ in the configuration of a single carbon. D-glucose and Dmannose are epimers. ALDOSES AND KETOSES Monosccharides – 2 types. Aldehydes aldoses. Ketones ketoses e.g. dihydroxyacetone (ketose) RIBOSE is a 5 carbon aldose – aldopentose. In RNA and ATP 2’ –OH isn’t found in DNA, prevents the formation of double helix due to steric hindrance. Phosphorylation of Carbohydrates • Key intermediates in energy generation and biosynthesis – transfer of phosphate group from ATP to glucose first step in glycolysis • Phosphorylation makes sugars anionic – can form strong electrostatic interactions with the active site of an enzyme and stops sugars crossing lipid bilayer membranes • Generation of reactive intermediates to form N- and O-glycosidic linkages Kinase – transfers a phosphoryl group. Phosphatase – removes a phosphoryl group. GLYCOSIDIC BONDS – formation of oligosaccharides. E.g. maltose is 2 glucose units joined by an Oglycosidic bond. Starch and glycogen have long chains of glucose molecules wuth alpha-1,4 linkages. Cellulose has beta-1,4 linkages. LACTOSE – disaccharide of galactose and glucose Cleaved by the enzyme lactase (belongs to beta-galactosidade enzyme family) in the gut to give galactose and glucose. o Lactose intolerance o Adults with insufficient lactase, lactose broken down by MOs in the colon producing lactic acid, methane and hydrogen. o Leads to gut distension, flatulence and diarrhoea. AMINO SUGARS A number of hexose Name the main types of chemical functional groups and their associated reactions in these molecules (for example; peptide bonds, glycosidic bonds) • Amino acids linked by peptide bonds – between COOH and NH2 groups • The peptide bond is a resonance hybrid between two contributing structures and has partial double bond character. • The amino-acid sequence of a peptide or protein is always specified from the amino terminus (N) to the carboxylate terminus (C). • Polypeptide chain – regular repeat backbone, variable side chains (R groups) GLYCOPROTEINS Proteins with oligosaccharide glycosylation N-linked through asparagine O-linked through serine/threonine Many of the proteins on the surface of animal cells have sugars attached to the amide nitrogen of asparagine residues or to the side chain oxygen of serine or threonine residues. SUGAR DERIVATIVES Carbonyl group of a monosaccharide can be reduced to a hydroxyl giving a sugar alcohol ot polyol. E.g. sorbitol, found in plants and berries, used a food additive The aldehyde can also be oxidised to give a carboxylic acid. Catalysed by glucose oxidase – H2O2 with peroxidase. PEPTIDE BOND FORMATION An addition/elimination reaction. Nucleophilic addition – nucleophilic attack of the carbonyl carbon by the lone pair of the amino group. Unstable intermediate results in the elimination of a water molecule. Delocalisation of the pi electron over the whole peptide bond, rather than just over C=O bond. Bond can be trans or cis. Trans is energetically favoured owing to lesser steric hinderance. DISULPHIDE BONDS – cross-links Form between 2 cysteine residues in proteins. Often found in extracellular proteins Increase stability of 3D structures. Insulin contains intra and inter-chain disulphide bonds. Synthesised in the pancreas as a single polypeptide chain – preproinsulin. Cross links generally reduce water solubility. ISOELECTRIC POINT Overall electric charge on a protein molecule depends upin the amino acid composition and the pH. pH at which a protein has no net electric charge = isoelectric point. Useful general rule, for an acid: pH > pKa [A-] dominates pH < pKa [HA] dominates Understand the biological function of the major families of biomolecules. AMINO ACIDS AS NEUROTRANSMITTES e.g. glutamate. Release leads to cation channels opening and membrane depolarisation. Glycine and gamma-aminobutyric acid (GABA) are inhibitory neurotransmitters. Lead to Cl- channels opening. Link chemical structure with function for selected biomolecules ABO BLOOD GROUP ANTIGENS – O-LINKED OLIGOSACCHARIDES H-antigen: precursor to ABO antigens. Modified by glycosyl transferases. A-antigen: extra GalNAc (oligosaccharide N-linked through asparagine. B-antigen: extra Gal (Galactose) DNA BASES – PURINES AND PYRIMIDINES 4 differen bases in DNA. Bicyclic – purines (A, G) Monocyclic – pyrimidines (C, T, U for RNA) NUCLEOSIDE: Nucleobase + ribose Adenine adenosine deoxyadenosine Guanine guanosine deoxyguanosine Cytosine cytidine deoxycytidine Uracil uridine Thymine thymidine deoxythymidine Deoxynucleosides have no 2’ OH NUCELOTIDE: base +ribose + phosphate Condensation of phosphate group on one nucleotide and the –OH atom of the next nucleotide = phosphodiester bridge linking nucleosides. 3 hydrogen bonds between C and G 2 hydrogen bonds between A and T OLIGONUCLEOTIDES Will bind to complementary sequences of single stranded DNA (or RNA) – HYBRIDISATION LIPIDS Water soluble, polar head group and non-polar tail. Variety of biological roles. E.g. energy storage (fat) and chemical messenger (hormones) SAPONIFIABLE (hydrolysable) – fatty acids, triacylglycerols, phosphoacylglycerols NON-SAPONIFIABLE (non-hydrolysable) – steroids, eicosanoids. Saponification refers to hydrolysis that converts and ester to a salt of carboxylic acid FATTY ACIDS COOH head-group and long hydrocarbon tail. C=C always cis Essential fatty acids, can’t be synthesised by mammals. TRIACYLGLYCEROLS – consist of 3 fatty acids. Chains esterified to a glycerol backbone. Hydrolysed during digestion: lipolysis Pancreatic lipases found in ileum (small intestine) Necessary so molecules small enough to be absorbed PHOSPHOACYLGLYCEROLS/GLYCEROPHOSPHOLIPIDS One fatty acids in triglyceride replaced by phosphoric acid Most abundant component of cell membranes Double tailed glycerophospholipid molecules form a bilayer CHOLESTROL Steroid: naturally occurring compound that contains a sterol derived from steroid and contains –OH group and 3 position Most common sterol, found in plasma membranes Transported and stored in the form of fatty acid esters Controls fluidity of membrane Precursor to other steroids EICOSANOIDS Polyunsaturated arachidonic acid, precursor to eicosanoid family Produced biosynthetically using COXm lipoxygenase and P450 enzymes Characteristic cyclic units in middle of hydrocarbon backbone Lecture 4 – Acids and Bases Compare and contrast the definitions of acids and bases described by the Brønsted-Lowry and Lewis acid-base theories. Bronsted-Lowry ACID PROTON DONOR BASE PROTON ACCEPTOR Lewis ACID can form new covalent bonds by accepting an electron pair. Electrophile BASE can form new covalent bond by donating an electron pair. Neutrophile Lewis bases are also Brønsted bases, many Lewis acids are not Brønsted acids. Define and describe pH, pKa, Ka and Kw and perform simple calculations relating to acids and bases pH The acidity of a solution is measure on a pH scale where pH = -log10[H+] [H+] = 10-pH For pure water, [H+] = 10-7 M pH < 7.0, then [H+] > [OH‐] solution is acidic pH > 7.0, then [H+] < [OH‐] solution is basic Logarithmic scale, small differences pH correspond to much larger differences in [H+] Cytosol – 7.2 Lysosome – 5.0 due to acid hydrolase/hydrolytic enzymes Mitochondria – 8 Strong acid: pH = -log10[H+] Strong base: Kw = [H+][OH-] = 10-14 M2 [H+] = 10-14/[OH-] pH = -log10[H+] Weak acid: pH = IONIC PRODUCT OF WATER: Ka = [H+][OH-]/[H2O] as conc of water is near constant, it’s not included so… Kw =[H+][OH-] Kw = 1x10-14 (roughly) For pure water: [H+] = [OH-] pKa HA H+ + A- pKa = -log10(Ka) Ka = [H+][A-]/[HA] Strong acids (hydrochloric) nearly completely dissociate. Large Ka and therefore a small pKa Weak acids (acetic) are mainly undissociated. Smaller Ka and therefore a larger pKa More dissociated, equilibrium shifts to the RHS = larger Ka = smaller pKa Describe what buffers are, give examples, and perform simple calculations relating to their use Buffer solution: a solution that resists changes in pH on the addition of small amounts of acid/base Weak acid and bases are able to buffer the pH of a solution over a range of 1 either side of pKa. HENDERSON-HASSELBALCH EQUATION HA H+ + A- Equation can be used to estimate the pH of a solution cantaining an acid and its conjugate base. When [A-] = [HA], pH = pKa Therefore half the acid is ionised when pH = pKa TITRATION: weak acid + strong base DIPROTIC ACIDS Amino acid glycine has 2 ionisable functional groups At neutral pH, amino acids are dipolar (zwitterion) and can act as an acid or a base. pI is isoelectric point, pH at which overall charge is zero. [H2A] = [A-] pI = (pKa1 + pKa2/2) POLYPROTIC ACIDS Phosphoric acid has 3 ionisable groups with different pKa values. Give examples of the effect of pH on functional groups in simple biomolecules, and reflect on how this affects their overall physiochemical properties Describe buffers found in biological systems, and the chemical species involved. o o o o o Properties of biological systems depend of pH. pH must be accurately controlled in any in vitro measurements Controlled using buffer solutions, pH remains essentially constant despite addition of small amounts of acid/base. ACID-BASE CONJUGATE PAIR Buffers useful within +/- 1 unit of pKa Tris(hydroxymethyl)aminomethane: o o o o Used as a buffer in biology/biochem Usually called Tris base 121 g mol-1 Conjugate acid pKa = 8.1 TITRATING BUFFERS o o o o Buffers normally made by titrating a weak acid with a strong base Measure pH with a pH meter, must be calibrated using reference solution. Solution of weak acid/base titrated with strong base/acid to obtain desired pH pH of buffers vary slightly with temperature. Some more than others. Lecture5 – Stereochemistry in Biomolecules Understand the different types of isomerism that can occur, and depict simple examples using standard notation. ISOMERISM: the different forms of molecules with the same formula can adopt as a consequence of the arrangement of their constituent atoms. Structural Isomers – same composition/molecular formula, differ in the order in which atoms are ordered together. Stereoisomers – same compositional/molecular formula and order in the bonding of their constituent atoms, differ in their 3D arrangement in space. o o o o Enantiomer (optical isomer), one that is non-superimposable on its mirror image. Must contain a chiral centre/stereocentre. Absolute configuration of a molecule is the description of the spatial arrangement of the stereocentres it contains. C atom with 4 separate bonds adopts a tetrahedral geometry. 4 different groups chiral carbon. Can exist in 2 isomeric forms Molecule with n stereocentres will have 2n stereoisomers. Named based on their… Optical activity: (+)- or d(-)- or l- dextrorotatory laevorotatory Configuration: CORN rule used to determine nomenclature and priority rules for each chiral centre. R- or S- according to orientation of the prioritised substituents. D- and L- related to the observation on the enantiomers of glyceraldehyde. NOT related to (+) or (-) nomenclature associated with optical activity. Naming compounds done by comparing stereocentre to that of D-glyceraldehyde (which is confusingly dextrorotatory) CORN RULE In an amino acid (not glycine), there are 4 different functional groups around central chiral carbon atom. Viewed with H group pointing away from viewer. CLOCKWISE D-amino acid ANTICLOCKWISE L-amino acid CAHN-INGOLD-PRELOG (CIP) PRIORITY RULES Assigns priority to groups at stereocentres Order of priority assigned based on atomic number (Z – number of protons) Where there are ties for priority based on Z, atoms at 2 bonds distance are then considered and so on until a ranking is determined. Stereocentre is viewed with group with lowest priority pointing away from the observer. Other groups are numbered by rank Order of the other 3 substituents is then deduces from the POV of the observer. a. R= clockwise b. S = anticlockwise CIP rules can also be used for the nomenclature of the groups around double bonds, which are in a fixed orientation. Groups attached to carbon atoms at each end of he double bond are assigned a priority according to atomic number. Z = on same side ‘Zusammen’ E = opposite sides ‘Entgegen’ Advantages: Doesn’t rely on historical comparisons or similarities to existing compounds. Done on easily determined ranking of atoms in a molecule Each stereocentre in a molecule can be easily described when more than one exists. Give a definition of the terms chiral centre, enantiomer, diastereoisomer. CHIRAL CENTRE: carbon atom with 4 different functional groups attached to it. Will rotate plane polarised light PPL. Able to produce enantiomers. ENANTIOMERS: Identical properties in a symmetric environment, apart from interaction with plant-polarised light PPL Each will rotate PPl in opposite direction to the same degree. Equal proportions of each enantiomer racemate. No effect on PPL rotation. DIASTEREOISOMERS Stereoisomers that differ in the absolute configuration of one or more of their stereocentres, are not mirror images of each other. Those differing in only one stereocentre are epimers. Unlike enantiomers, they can have different physical and chemical properties. Be able to work out the absolute configuration of simple biologically-relevant molecules from their skeletal representation. Give examples of chiral biomolecules, and indicate the relevance of chirality in biochemical interactions. Many molecules in a biological environment are chiral, and therefore will have a differential interaction with each enantiomer. Enantiospecific recognition by proteins Proteins in eukaryotes are comprised of L-amino acids only. Thalidomide Drug used in the 1950s as a sedative. Withdrawn after adverse side effects were observed Has a single stereocentre. S- enantiomer found to treat morning sickness R- enantiomer is a teratogen (causes birth defects) Be able to interpret Fischer projections of simple molecules such as sugars in terms of the stereochemical information they contain. PROJECTIONS There are numerous ways to depict the structural formula of molecules. Some give no spatial/geometric information – e.g. Lewis structures Several can be used to denote 3D arrangements (and therefore stereochemistry) in 2D – Fischer projection, Haworth projection, Newman projection. FISCHER PROJECTIONS Typically used to depict sugars and amino acids Stereochemistry can be compared easily. Needed for sugars as there are a lot of stereocentres Carbon chain conventionally drawn vertically with bonds to other atoms horizontal. Vertical bonds represent bonds pointing away from observer Horizontal bonds represent bonds pointing towards observer Follow the format of D-glyceraldehyde HAWORTH PROJECTIONS Ring structures are more easily interpreted using a Haworth projection Depiction of carbon and hydrogen atoms in ring is similar to normal skeletal formula. Ring depicted with thicker lines towards observer, thin lines away. INTERCONVERSION OF FISCHER AND HAWORTH PROJECTIONS (for sugars) STEPS: 1. 2. 3. 4. Draw Fischer projection using conventional approach Rotate diagram 90 degrees clockwise Start indicating the ring curvature, indicate the –CH2OH is moving backwards Ensure to keep stereochemistry at C5 constant and rotate about C4-C5 bond to position the –OH to form the ring. 5. Close the ring. Lecture 6 – Organic Reactions Understand the principles of thermodynamics and kinetics. Thermodynamics answers the questions: 1. Is the reaction favourable? Exo or endothermic? 2. In which direction does the reaction go? Is equilibrium shifted towards the products of reactants? 3. To what extent is the reaction completed? Calculate the free energy of reactions and predict their outcome Potential energy of a molecules expressed as change in free energy ∆Gº (Gibbs) under standard state conditions ∆Gº = ∆Hº - T∆Sº T = temperature H = enthalpy (bond energies) S = entropy (degree of disorder) o o o Can determine the relative importance of enthalpy and entropy for a given reaction, most reactions will have little entropy change. As T increases, entropy increases. Favourable reactions ∆H<0, ∆S>0 Understand how to interpret reaction profiles for catalysed and non-catalysed reactions CATALYSTS: altered mechanism that lowers activation energy. o Modified the reaction pathway or stabilize the transition state. o Position of equilibrium not affected by approaches equilibrium more rapidly. Enzymes – biological catalysts o o All biological reactions are catalysed by enzymes Active sites of enzymes are designed to stabilise the transition state, decrease activation energy required. Understand the concepts of activation energy, transition states, intermediates and rates of reactions. Activation Energy, Ea, of a reaction is the energy in excess of that possessed by the ground state that is needed for a reaction to proceed. TRANSITION STATE SN2 reactions – species of highest energy along reaction pathway, transient existence, highly unstable, breaking of old and formation of new bonds. Simple one-step SN2 reaction carried out. Only occurs at energy maximum, and cannot be isolated. MULTI-STEP REACTION AND REACTIVE INTERMEDIATES o SN1 reactions, real time existence, exist at bottom of potential energy well and must overcome and energy barrier for reaction to proceed. o Highest activation energy for a given transition state is the rate limiting/determining step – control overall rate of reaction. o Intermediate occurs at local energy minimum and can be detected and potentially isolated. Apply curly arrows to describe the movement of electrons and bonding. BOND MAKING RELEASES ENERGY BOND FORMING TAKES IN ENERGY Generates Radicals – electron pair of sigma bond shared between A and B. Very reactive species containing unpaired electron Electron pair in bond assigned to same fragment, direction of electron flow determined by electronegativity. Heterolytic cleavage of C-H bond Carbanion and proton formed or carbocation and hydride anion. Electron Pushing o Curly arrows represent movement of an electron pair. o Total number of electrons doesn’t change o Half headed arrow represents a single electron being transferred. Resonance…. Electrons move away from the negative charge, and move towards positive. Understand the terms nucleophile and electrophile, and recall why these concepts are important in chemical reactions. ELECTROPHILE: electron pair acceptor. Often carry a partial positive charge. E.g. H+ NUCLEOPHILE: electron pair donor. Sometimes carrying a partial negative charge. E.g. Cl-. Double and triple bonds are nucleophilic. e.g. Acid-base reaction. Brønsted-Lowry Nucleophilic substitution – nucleophile in and leaving group out. Nucleophilic addition - Electrophilic addition – double bond acting as a nucleophile, formation of a carbocation. Lecture 7 – Reaction Mechanisms Reaction mechanism – a detailed and sequential description of the changed in structure and bonding that take place in the course of a reaction. o o o o o o o is it a concerted or multistep reaction? representation of plausible electron reorganization description of the transition state identification of any intermediate species stereochemistry of the products kinetic analysis (which is the rate-limiting step?) molecularity = the number of species involved in the rate limiting step Describe the fundamentals of major reaction mechanisms which include addition, elimination, substitution (SN1/SN2), condensation and oxidation-reduction. ELIMINATION o Number of s-bonds in the substrate molecule decreases and one or more pi bonds are formed. ADDITION o Number of s-bonds in substrate molecule increases, usually at expense of pi bonds. SUBSTITUTION o The replacement of an atom or group (Y) by another atom or group (Z) REARRANGEMNT o Generates an isomer, and generally the number of bonds doesn’t change. SN2 – biomolecular nucleophilic substitution Rate = k[A][B] Rate limiting step involves both species. Reaction mechanism is concerted, single step, reaction proceeds from reactant to product without forming a detectable intermediate. Concerted bond making and bond breaking occur simultaneously. No intermediated formed. Bond formation occurs on opposite side of bond break, causes an inversion of configuration. Changed enantiomer R to S. Back-side displacement, strongly favoured over front-side displacement as it would put both electron rich atoms in close proximity Steric Hindrance of Substrate – transition state is crowded by bulky substituents on carbon, making it less stable. Reactivity: CH3Br > CH3CH2Br > (CH3)3CHBr > (CH3)3Br Primary > secondary > tertiary. SN1 – unimolecular nucleophilic substitution Rate = k [A] Only one molecule involved in the rate-limiting step. 2 STEP REACTION C-X bond completely broken before the formation of a new C-Nuc bond. Step 1 is the rate determining step. Step 2 is the fast step and doesn’t affect the overall reaction rate. 2 steps – 2 transition states. Reaction rate proportional to the concentration of (CH3)3Br as first step only involves (CH3)Br Stability of carbocations increasing with the number of substitutents. TERTIARY > SECONDARY > PRIMARY > METHYL o o Attack by nucleophile on either side of planar carbocation is equally likely. Reactive carbon is a chiral centre. The product is a pair of enantiomers, racemic mixture. ELIMINATION REACTIONS Can compete with substitution of an incoming nucleophile for X-E1 unimolecular, E2 bimolecular. E1 - unimolecular: rate of reaction only dependant on haloalkane, same as first step of SN1. Formation of carbenium ion. Second step fast, where Nuc- extracts H+ from carbenium ion intermediate. Factors that affect whether Nuc- adds as in SN1 or E1 occurs – abstraction of H+ facilitated by a strong base that is poor Nuc- or steric factors in substrate may favour E1 E2 Biomolecular rate determining step Elimination of HX from haloalkane Rate of reaction = k[haloalkane][base] Reaction profile Transition state: base begins to form bonds with proton for elimination and simultaneously the C-H bond begins to break. Pi-Pi component of C=C begins to form and C-Br bond begins to break Competition with SN2 if Nuc- is a strong base. E2 favoured if Nuc- is sterically bulky. Have an understanding of the role of these mechanisms in important biological reactions (e.g. peptide and glycosidic bond formation). OXIDATION-REDUCTION Very common reactions in biology. 2 compounds that accept or donate hydrogen atoms and/or electron in a biological context: NAD/NADH – nicotinamide adenine dinucleotide o Major redox agent in biological reactions. o Nicotinamide ring acts as hydride donor (or acceptor in NAD) o Donates of hydride in reduction FAD/FADH2 – Flavin Adenine Dinucleotide o Ring system acts as a hydrogen/electron acceptor/donor o In reduction, FADH2 donates 2 H+ and 2eo In oxidation it accepts 2H+ Peptide bond formation 1. Nucleophilic addition 2. Elimination of water from unstable intermediate. Condensation Nucleic acid synthesis Formation of phosphodiester bonds in nucleic acids 1. Nucleophilic addition. 2. Elimination of pyrophosphate from unstable intermediate. Lecture 8 – Analytical Approaches for Structural Elucidation of Biomolecules Pt1 Describe the analytical tools for structural elucidation of both small and large molecules. Understand the information obtained, and the advantages and limitations of each technique. Outline the principles of UV/Vis, Fluorescence, IR spectroscopy, and X-ray crystallography. X-RAY CRYSTALLOGRAPHY o o X-rays passed through crystals and diffraction patterns analysed to calculate 3D structure. o When waves pass through gap, they spread out diffraction o 2 gaps, wave spread from each – meet and cause interference (contructive or destructive) Pattern shows the spatial location of the atom centres in a molecule. ULTRA VIOLET AND VISIBLE SPECTROSCOPY o When sample molecules exposed to light, some of light energy absorbes as electron is promoted to a higher energy orbital. When energy matches the possible electronic transition. CHROMOPHORE: a chemical group that absorbs light and so imparts colour to a molecule. Light absorption A is the absorbance or optical density of the sample BEER-LAMBERT LAW A – absorbance (no units) Ε – molar extinction coefficient (M-1cm-1) C – concentration (M) D – path length (cm, cuvette size) UV spectrophotometers operate down to 200nm o Ordinary single and double bonds absorb at wavelengths less than this. o Conjugated bonds absorb at longer wavelengths (bathochromic) Conjugated bond – a system where 2 multiple bonds are separated by a single bond. Show awareness of the use of analytical techniques in determining the structure of molecules. Uses of UV/VIS spectrophotometery Spectrophotometry is used to measure the concentration of chromophores in solution. Proteins and nucleic acids absorb in the UV and can be detected directly. Some enzyme reactions involve colour changes that can be followed directly. Other measurements involve using a chemical reaction (often enzyme catalysed) that produces a coloured product. Amino acids only tryptophan and tyrosine absorb at 280. Nucleic acids 260nm. Absorbance depends on environment; double stranded absorbs about 30% less strongly than single stranded DNA. NAD+ to NADH Spectrophotometry can be used to monitor the course of a chemical reaction, Lactic acid fermentation – reduction of pyruvate to lactate catalysed by the enzyme lactate dehydrogenase. Uses co-factor NADH Monitory at 340nm – NADH absorbs but NAD+ doesn’t. (practical) FLUORESCENCE o o o o Instead of losing energy if an excited state as heat, a molecule can lose energy by the emission of light: fluorescence A chemical group/molecule that fluoresces is called a fluorophore – amino acids, flavins, vitamin A and NADH. Fluorophores can be attached to other molecules to act as visual probes. Fluorescence can be used to determine sample concentration and to follow the course of a reaction. Feature of Fluorescence o The emitted photons will be of a lower energy – longer wavelength – than those absorbed. o Absorption spectrum often mirrors the emission spectrum o Fluorescence is sensitive to the environment and the fluorophore – much more so than absorbance o Collision of a fluorophore with solvent molecules can lead to the decay of the excited sate without light emission – quenching. o Intense illumination can destroy flurorophore – photobleaching USES Visualisation of DNA in gels and when sequencing – ethidium bromide in agarose gels. Emits orange light. Immunofluorescence for seeing protein in cells – cells stained with fluorescently labelled antibodies, examined under fluorescent microscope FISH for labelling specific DNA regions – fluorescently labelled complementary DNA probe FRET (fluorescence resonance energy transfer) GFP (green fluorescent protein) for in vivo visualisation of proteins – from jellyfish o GFP chimeras used to localize cells expressing protein of interest and reveal subcellular localization o GFP variants generated that emit light a different wavelengths o Can label more than one protein using different colours for each o FRET is CFP (cyan) and TFP (yellow) combined. Chimera: a single organism (usually an animal) that is composed of two or more different populations of genetically distinct cells that originated from different zygotes involved in sexual reproduction. INFRA-RED SPECTROPHOTOMETRY IR is used to differentiate between bond types and functional groups o o o o o Vibrational motions, characteristic of the component atoms of a molecule All organic compounds will absorb IR that corresponds in energy to these vibrations Applied to samples in all phases For vibration to be detected, IR spectrum must occur within a change in dipole moment. Symmetric stretch no dipole moment no IR response. Lecture 8 Summary: Absorbance and fluorescence are both the result of the interaction of light with the electrons of molecules. Electrons are excited from the highest occupied molecular orbital to the lowest unoccupied orbital. Light energy is ultimately lost as longer wavelength electro-magnetic radiation, but in absorbance, this is heat radiation, whereas in fluorescence it is mostly light. Both fluorescence and absorbance are common in compounds with conjugated double bonds. Absorption can be used to quantifying protein and nucleic acid concentrations – using the Beer-Lambert law. Absorption is used with enzyme coupled reactions. Fluorescence used for visualizing proteins and nucleic acids. GFP can be used to visualize proteins in living cells Lecture 9 – Analytical Approaches for Structural Elucidation of Biomolecules Pt 2 Describe analytical tools for structural elucidation of both small and large molecules Understand the information obtained, and the advantages and limitations of each technique. Outline the principles of nuclear magnetic resonance spectroscopy, chromatography and mass spectrometry. NMR nuclear magnetic resonance o Generates spectra that describe the chemical environment of nuclei in a sample. Chemical structure Interactions Bulk properties Diffusion Compartmentation o Concentrates on1H o Very reproducible/robust/non-destructive platform. When a magnetic field is applied to the same, protons align either with or against it. Nuclei in different chemical environments will experience different magnetic fields – different frequencies. Fourier Transformation (FT) – mathematical transformation of time dependent signal, following a pulse loss of energy as nuclei return to ground state, detected as function of time. Spin Coupling Area under peak (integral) directly proportional to number of nuclei giving resonance. N+1 rule to determine peak splitting. Limitations: relatively insensitive, limited detection. Increasing field strength is improving sensitivity. MASS SPECTROMETRY Characterises components by: Mass-to-charge ratio (m/z) Fragmentation pattern (MS/MS) Used for structural elucidation Masses of fragments give partial structural information Tryptic digest can also be used – information on resulting peptides acquired A variety of spectrometers are available Q-ToF and FTMS provide very high m/z accuracy/precision that can allow deduction of the empirical formula MS coupled to chromatography Separates components Helps deconvolve complex mixtures Retention times are usually very stable. Provide an additional characteristic for identification Very sensitive (picomol), to detect minor components Used extensively with NMR for rapid structure elucidation of unknowns MS IONISATION TECHNIQUES o Measure the mass of a molecule once converted to a gas-phase ion, o Imparts and electrical charge to molecules and converts the resultant flux of electrically charged ions in to a proportional electrical current. Techniques: Laser ablation of components dissolved in a matrix Interaction with an energized particle or electron Electrospray, eluent (substance used as a solvent in separating materials in elution) from a liquid chromatograph receives a high voltage. o Sample heated until vapourises, bombarded with high energy electrons ADVANTAGES OF MS Rapid method for structural info – high sensitivity Separates nioflui components –used in metabolism/pharmacokinetic studies Easily automated Information rich datasets DISADVANTAGES OF MS Infusion MS generates very complex, overlapping spectra LC/MS more complicated and generates very large data sets HPLC – high performance liquid chromatography Solvent (mobile phase) High pressure pump HPLC column, chromatographic packing materials – separates metabolites as some retain on column and others elute (the process of extracting one material from another by washing with a solvent. Detector typically UV – absorbance or fluorescence. POLARITY, ELECTRICAL CHARGE, MOLECULAR SIZE o Compounds with similar polarity to stationary phase will be retained o Compounds whose polarity is similar to that of the mobile phase will be eluted. Show awareness of the use of analytical techniques in determining the structure of molecules. NMR is applied to macromolecule in structural genomics