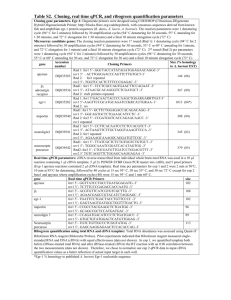
Table S1 : Detailed overview of strategies used to assemble the
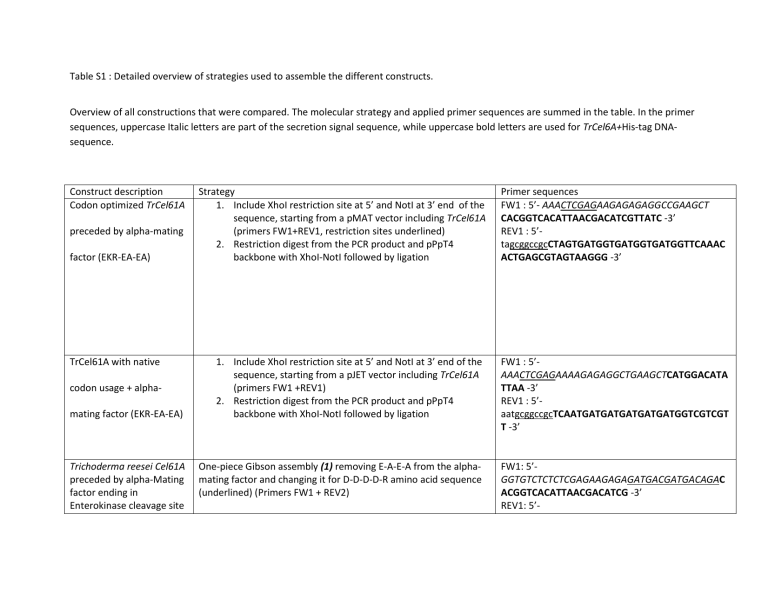
Table S1 : Detailed overview of strategies used to assemble the different constructs.
Overview of all constructions that were compared. The molecular strategy and applied primer sequences are summed in the table. In the primer sequences, uppercase Italic letters are part of the secretion signal sequence, while uppercase bold letters are used for TrCel6A+His-tag DNAsequence.
Construct description
Codon optimized TrCel61A preceded by alpha-mating factor (EKR-EA-EA)
Strategy
1.
Include XhoI restriction site at 5’ and NotI at 3’ end of the sequence, starting from a pMAT vector including TrCel61A
(primers FW1+REV1, restriction sites underlined)
2.
Restriction digest from the PCR product and pPpT4 backbone with XhoI-NotI followed by ligation
Primer sequences
FW1 : 5’- AAACTCGAGAAGAGAGAGGCCGAAGCT
CACGGTCACATTAACGACATCGTTATC -3’
REV1 : 5’tagcggccgcCTAGTGATGGTGATGGTGATGGTTCAAAC
ACTGAGCGTAGTAAGGG -3’
TrCel61A with native codon usage + alphamating factor (EKR-EA-EA)
1.
Include XhoI restriction site at 5’ and NotI at 3’ end of the sequence, starting from a pJET vector including TrCel61A
(primers FW1 +REV1)
2.
Restriction digest from the PCR product and pPpT4 backbone with XhoI-NotI followed by ligation
Trichoderma reesei Cel61A preceded by alpha-Mating factor ending in
Enterokinase cleavage site
One-piece Gibson assembly (1) removing E-A-E-A from the alphamating factor and changing it for D-D-D-D-R amino acid sequence
(underlined) (Primers FW1 + REV2)
FW1 : 5’-
AAACTCGAGAAAAGAGAGGCTGAAGCTCATGGACATA
TTAA -3’
REV1 : 5’- aatgcggccgcTCAATGATGATGATGATGATGGTCGTCGT
T -3’
FW1: 5’-
GGTGTCTCTCTCGAGAAGAGAGATGACGATGACAGAC
ACGGTCACATTAACGACATCG -3’
REV1: 5’-
(EKR-DDDDR-) CGATGTCGTTAATGTGACCGTGTCTGTCATCGTCATCTC
TCTTCTCGAGAGAGACACC -3’
TrCel61A with native codon usage +
Method as described by Sanchis et al (2008) (2) to remove aminoacids E-A-E-A at the 3’ end of the alpha-mating factor starting from the complete vector with alpha-mating factor ending in EKR-E-A-E-
A (Primers FW1 +REV1)
FW1: 5’-
GGTGTCTCTCTCGAGAAGAGACACGGTCACATTAACGA
CATCG -3’
REV1: 5’- gaagaggagtgggaaatacc -3’ alpha- mating factor (EKR)
DDDK protein native secretion signal
Phanerochaete
chrysosporium GH61D
1.
Amplify TrCel61A with the DDDK-protein secretion signal in front via PCR amplification and including the sequence into the primer (primers FW1+REV1)
2.
Gibson assembly (1) to insert gene and secretion signal into pPpT4 vector (insert amplification primers: FW2
+REV1, backbone primers: FW3 + REV2)
Gibson assembly (1) to exchange TrCel61A sequence to pPpT4 vector containing already Phanerochaete chrysosporium GH61D secretion signal (insert amplification primers: FW1 +REV1, backbone primers: FW2 + REV2)
FW1: 5’-
CTACTTTGGCTTCCATTGCTGTTGCTCACGGTCACATTA
ACGACATCGTTATC -3’
REV1: 5’- cttgagcggccgcctAGTGATGGTGATGGTGATGGTTCAAA
CACTGAGCGTAGTAAGG -3’
FW2: 5’-
GTTTAACTTGAAGACTATTTTGATTTCTACTTTGGCTTCC
ATTGCTGTTGCT -3’
FW3: 5’-
CCTTACTACGCTCAGTGTTTGAACCATCACCATCACCAT
CACTAGgcggccgctcaag -3’
REV2: 5’-
CAAAATAGTCTTCAAGTTAAACATcgtttcggaattctttcaat aattag -3’
FW1: 5’-
GTTGTCTCTGCTCCATTTGTCTTGGGTCACGGTCACATT
AACGACATCG -3’
REV1: 5’- ccgctcAATGATGATGATGATGATGGTTCAAACACTGAG
CGTAGTAAGG -3’
Trichoderma reesei Cel61A preceded by its native secretion signal sequence
1.
Precede TrCel61A by its native secretion signal sequence via PCR primers and include EcoRI restriction site at the 5’ and NotI site at 3’ end of the sequence (primers FW1 +
REV1, restriction sites underlined)
2.
Restriction digest from the PCR product and pPpT4 backbone with EcoRI-NotI followed by ligation
FW2: 5’-
CCTTACTACGCTCAGTGTTTGAACCATCATCATCATCAT
CATTGAgcgg -3’
REV2: 5’-
CGATGTCGTTAATGTGACCGTGACCCAAGACAAATGGA
GCAGAGACAAC -3’
FW1 : 5’- aaagaattccgaaacgATGATTCAAAAATTGTCTAACTTACT
TGTTACTGCTTTGGCAGTTGCTACTGGTGTTGTGGGAC
ACGGTCACATTAACGACATCGTTATCAAC - 3’
REV1 : 5’tagcggccgcCTAGTGATGGTGATGGTGATGGTTCAAAC
ACTGAGCGTAGTAAGGG -3’
1. D.G. Gibson, L. Young, R. Chuang, et al. (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. 6, 12–16.
2. J. Sanchis, L. Fernández, J.D. Carballeira, et al. (2008) Improved PCR method for the creation of saturation mutagenesis libraries in directed evolution: application to difficult-to-amplify templates. Applied microbiology and biotechnology. 81, 387–97.









![Introduction to Second Law (contd.) [Lecture 4].](http://s2.studylib.net/store/data/005767600_1-250da23339ac97bb85a82c10f66d9a9f-300x300.png)