4.1 Simple Linear Regression R code
advertisement

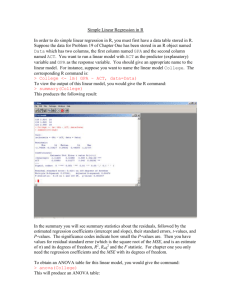
Simple Linear Regression R code Suppose we have $100 in the bank, and we work 40 hours at $10 per hour. At the end of the week, we have a total of 100 + 40*10 = 100 + 400 = 500 dollars. We can create a graph of how much money we have after each hour of work. hours.worked = 0:10 dollars = 100 + 10 * hours.worked plot(hours.worked, dollars, ylim=c(0,200)) We can draw a straight line through the points using the abline() function. abline(100,10) The slope of the line is 10, because we earn 10 dollars per hour. The intercept of the line (when x = 0) is 100, because at time 0 we have 100 dollars. So the equation for the regression line is y = 100 + 10x dollars = 100 + 10 * hours.worked The intercept (100) and slope (10) of the line are called coefficients. Perhaps we have a job that pays tips, rather than a fixed hourly wage. Suppose we get these tips for working 1 to 8 hours. hours.worked = c(1,1,2,2,3,3,4,4,5,5,6,6,7,7,8,8) tips=c(11,12,19,22,29,33,41,44,54,56,59,60,71,74,82,84) lm.tips = lm(tips ~ hours.worked) summary(lm.tips) Call: lm(formula = tips ~ hours.worked) Residuals: Min 1Q -3.2679 -1.6830 Median 0.2232 3Q 1.4226 Max 3.9524 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 0.9464 1.1675 0.811 0.431 hours.worked 10.2202 0.2312 44.205 <2e-16 *** plot(hours.worked,tips, xlab="Hours worked", ylab="Tips", ylim=c(0,100)) abline(lm.tips) We would like to know the average tips per hour worked, which is the slope of the line through the points. We use linear regression to calculate the slope, to learn the average tips per hour. Estimating coefficients in simple linear regression We estimate the coefficients (slope and intercept) using the method of least squares. Least squares regression finds the values of the coefficients that minimize the sum of the squared distance of all the points from the regression line. That is, the coefficients fit the line that minimizes the sum of the squared residuals. Use lm() to fit the regression For this example, we'll use a data set from Michael Crawley's textbook "The R Book", which is a very comprehensive description of statistical analyses using R. I've found it to be a great reference. The data set "tannin.txt" gives the growth of caterpillars on diets containing different amount of tannin. This is from Chapter 10, Regression, of Crawley's text, where he calls the file "regression.txt". setwd("C:/Users/Walker/Desktop/UCSD Biom 285/Data") tannin.data= read.table("tannin.txt", header=TRUE) > tannin.data growth tannin 1 12 0 2 10 1 3 8 2 4 11 3 5 6 4 6 7 5 7 2 6 8 3 7 9 3 8 with(tannin.data, plot(tannin, growth)) # fit regression line lm.tannin = lm(growth ~ tannin, data= tannin.data) This command tells R to regress growth on the variable tannin. The output of the lm() function is a model object, which we named lm.tannin. We can get some information about the model this way: > lm.tannin Call: lm(formula = growth ~ tannin, data = tannin.data) Coefficients: (Intercept) 11.756 tannin -1.217 The lm.tannin model includes the coefficients: The intercept is 11.756 The slope (coefficient for tannin) is -1.217. So the linear model is growth = 11.756 -1.217 * tannin This model indicates that for every one unit of tannin, growth decrease by 1.217 units. We can pass the lm.tannin object to other functions to create plots or extract information. The abline function will add the regression line to the scatterplot we created earlier: abline(lm.tannin) We can get more information about the model: summary(lm.tannin) > summary(lm.tannin) Call: lm(formula = growth ~ tannin, data = tannin.data) Residuals: Min 1Q Median -2.4556 -0.8889 -0.2389 3Q 0.9778 Max 2.8944 Coefficients: Estimate Std. Error t value (Intercept) 11.7556 1.0408 11.295 tannin -1.2167 0.2186 -5.565 --Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 Pr(>|t|) 9.54e-06 *** 0.000846 *** ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 Residual standard error: 1.693 on 7 degrees of freedom Multiple R-squared: 0.8157, Adjusted R-squared: 0.7893 F-statistic: 30.97 on 1 and 7 DF, p-value: 0.000846 Here's the meaning of the output: Call: lm(formula = growth ~ tannin, data = tannin.data) This is the formula that R used to perform the linear regression. We'll look at residuals in more detail later. Coefficients: Estimate Std. Error t value (Intercept) 11.7556 1.0408 11.295 tannin -1.2167 0.2186 -5.565 --Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 Pr(>|t|) 9.54e-06 *** 0.000846 *** ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1 The coefficients are usually the thing we are most interested in. In particular, we want to know if the coefficients are significantly different from zero. This section of the output gives us the coefficients that we saw above, but in addition gives us their standard error, t-value, and the associated p-value. The p-value tells us if the coefficient is significantly different from zero. For tannin, p-value = 0.000846, so we reject the null hypothesis that the true slope of the line is zero, and conclude that growth is significantly related to tannin. Residual standard error: 1.693 on 7 degrees of freedom Multiple R-squared: 0.8157, Adjusted R-squared: 0.7893 F-statistic: 30.97 on 1 and 7 DF, p-value: 0.000846 The residual standard error and R-square statistics tell us how well the line fits the data. Multiple R-squared takes values between 0 and 1, with 1 being the best possible. The multiple R-squared tells us how much of the variability in y (growth) is explained by x (tannin). We'll look at these more later. The F statistic is a test that the overall model is significant. We're usually more interested in whether or not individual coefficients are significant. The lm object, lm.tannin, also has the fitted values and the residuals. A fitted value is the value of y on the regression line corresponding to a particular value of x. It is the predicted value of y for a given x. Let's calculate and plot the predicted value of growth for tannin = 2 and tannin =6. Recall that the linear model was growth = 11.756 -1.217 * tannin growth = 11.756 -1.217 * 2 = 9.322 growth = 11.756 -1.217 * 6 = 4.454 abline(9.322,0, col=2) abline(4.454,0, col=3) Here are the fitted values: fitted(lm.tannin) > fitted(lm.tannin) 1 2 3 4 5 6 7 8 11.755556 10.538889 9.322222 8.105556 6.888889 5.672222 4.455556 3.238889 9 2.022222 The residuals are the difference between the fitted (predicted) value and the actual value resid(lm.tannin) 1 2 3 4 > resid(lm.tannin) 1 2 3 4 5 6 7 0.2444444 -0.5388889 -1.3222222 2.8944444 -0.8888889 1.3277778 -2.4555556 8 9 -0.2388889 0.9777778 You can get just the residual plot: Scatterplot with the fitted regression line (use the lines() function as alternative to abline) with(tannin.data, plot(tannin, growth)) with(tannin.data, lines(tannin,fitted(lm.tannin))) Add the residuals to the plot with(tannin.data, segments(tannin,fitted(lm.tannin), tannin, growth)) Testing the model assumptions The linear model makes several assumptions about the data that we should check before performing inference. R has several plots and tests to help. The residuals should be roughly normally distributed, and not show any pattern. with(tannin.data, plot(tannin, resid(lm.tannin))) qqnorm(resid(lm.tannin)) The residuals appear to be normal and don't show a pattern. Here's an example of residuals that show a pattern that indicates a problem. We'll generate data with a quadratic term to create curvature. Use a scatter plot to look for linearity vs. non-linear response x = c(1:10) y = x - (x-5)^2 + rnorm(10,sd=2) plot(x,y) This scatterplot shows clear curvature, so a simple linear model is not appropriate. We could add an x-squared (quadratic) term to the model to account for the curvature. The tannin versus growth plot doesn't show curvature, so a linear model is OK.