answers
advertisement
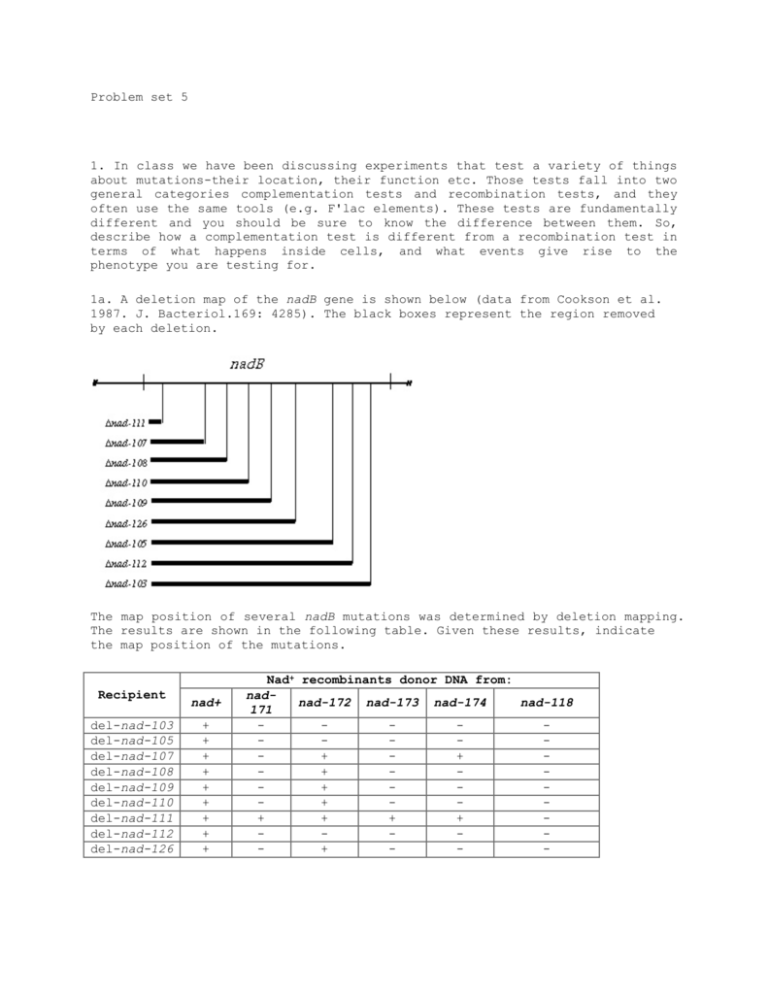
Problem set 5 1. In class we have been discussing experiments that test a variety of things about mutations-their location, their function etc. Those tests fall into two general categories complementation tests and recombination tests, and they often use the same tools (e.g. F'lac elements). These tests are fundamentally different and you should be sure to know the difference between them. So, describe how a complementation test is different from a recombination test in terms of what happens inside cells, and what events give rise to the phenotype you are testing for. 1a. A deletion map of the nadB gene is shown below (data from Cookson et al. 1987. J. Bacteriol.169: 4285). The black boxes represent the region removed by each deletion. The map position of several nadB mutations was determined by deletion mapping. The results are shown in the following table. Given these results, indicate the map position of the mutations. Recipient del-nad-103 del-nad-105 del-nad-107 del-nad-108 del-nad-109 del-nad-110 del-nad-111 del-nad-112 del-nad-126 nad+ + + + + + + + + + Nad+ recombinants donor DNA from: nadnad-172 nad-173 nad-174 nad-118 171 + + + + + + + + + + - 2. Proteins encoded by the put operon (put=proline utilization) allow cells to use proline as a sole carbon or nitrogen source. A deletion map of part of the put operon is shown below. (The open bar shows the region deleted in the indicated mutant.) Imagine that you have isolated a new mutation put127 that cannot use proline as a carbon or nitrogen source. Reversion analysis shows that put127 reverts to w.t. at a frequency of about 10-7, indicating that it is probably a point mutation of some sort. You bring the put region with the put127 allele into each of the deletion strains and select for recombinants that can use proline to grow. If you fail to find any, where does the put127 mutation likely map? It likely maps to region iii, because this area is missing in all the deletion strains 3. A new putP mutation was mapped against a set of putP deletion mutations. The region removed by each deletion mutations is indicated by an open box below the putP map. The results indicating whether or not recombinants were obtained are shown to the right of each deletion. Based on these results, where does the new putP mutation map with respect to the deletion intervals? 4. What hypothesis was being tested in the PAJAMO experiment and what was the result? 5. A series of five temperature sensitive (Ts) mutants of phage T4 were isolated and analyzed by complementation tests. The tests were performed by putting a mixture of the two phage mutants to be tested (about 106 of each) onto a lawn of bacteria at 40°C. The results are summarized below: (+ = complete lysis; - = no lysis or only a few plaques in the spot) a. How many complementation groups are represented by these mutants? There are 3 complemention groups b. Which mutations fall in each complementation group? The groups contain: “1 and 4”, “2” and “3 and 5” 6. There are two classes of lacI mutations which are dominant. Name the two classes and explain why each type of mutant is dominant over w.t. LacI. 7. Using the codon chart below, find all the codons that can mutate to a stop codon with a single base change. List the codons, and the amino acids that they encode. Note: some may be able to mutate to more than one type of nonsense codon. (Note: you should memorize the 3 stop codons)