3.1 T-tests R code
advertisement

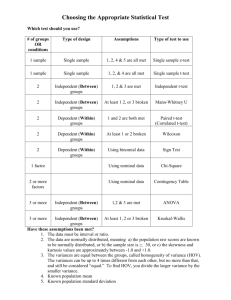
T-tests R code T-test for two independent samples We will test the ability of a drug to kill bacteria. The response is the count of bacteria colony forming units (CFU) x106 per milliliter of blood. Specifically, we will use a two-sample t-test to test the null hypothesis: mean bacteria count in the drug group = mean bacteria count in placebo group The standard version of the two-sample t-test assumes the following. the samples are normally distributed the two samples have equal variance The standard version of the two-sample t-test will usually give valid results if the data do not completely meet these assumptions. However, if the samples are strongly nonnormal, or have greatly different variance, we should consider alternative analyses, such as log transforming the data to get a normal distribution, or using non-parametric Wilcoxon rank tests described later. From our experiment, we collect the following data. drug.bacteria.counts =c(0.7, 0.9,1.0, 1.2, 2.2, 2.5, 3.6, 3.9, 4.2, 4.5, 4.5, 5.6, 5.9, 6.1) placebo.bacteria.counts=c(2.0,2.3, 2.3, 3.9, 4.1, 4.4, 5.3, 5.7, 6.3, 6.4, 7.2, 7.7, 8.0) We'll use boxplots and qqnorm plots to see if the counts deviate greatly from normality, or if there are outliers. boxplot(drug.bacteria.counts ) boxplot(placebo.bacteria.counts ) qqnorm(drug.bacteria.counts ) qqline(drug.bacteria.counts) qqnorm(placebo.bacteria.counts ) qqline(placebo.bacteria.counts ) The boxplots and qqnorm plots do not show significant non-normality or outliers. What about the assumption of equal variance? We'll calculate the standard deviation of each of the treatment groups using the sd() function. sd(drug.bacteria.counts ) sd(placebo.bacteria.counts ) > sd(drug.bacteria.counts ) [1] 1.923024 > sd(placebo.bacteria.counts ) [1] 2.067452 The standard deviations of the two treatment groups are similar. In this case, the boxplots and qqnorm plots do not show significant non-normality or outliers, and the standard deviations of the two treatment groups are similar. So we'll use the standard version of the t-test assuming equal variance. We can do a t-test directly with these values, using the t.test() function. t.test(drug.bacteria.counts, placebo.bacteria.counts, var.equal=TRUE) Two Sample t-test data: drug.bacteria.counts and placebo.bacteria.counts t = -2.2182, df = 25, p-value = 0.03586 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: -3.2847862 -0.1218072 sample estimates: mean of x mean of y 3.342857 5.046154 Conclusion: We reject the null hypothesis. Bacterial count was significantly less in the drug group than in the placebo group (p = 0.03586). The 95% confidence interval for the true value of the difference of the means ranges from -3.2847862 to -0.1218072. Because the 95% confidence interval does not include zero, we reject the null hypothesis that the difference between the two groups is zero. T-test using biomarker data set. setwd("C:/Users/Walker/Desktop/UCSD Biom 285/Data") biomarker.data= read.csv("biomarkers.csv", header=TRUE) biomarker.data > biomarker.data ID Sex Age Disease Biomarker.1 Biomarker.2 Biomarker.3 1 1 Female 30 Case 138 137 79 2 2 Female 30 Control 141 143 93 3 3 Female 40 Case 134 148 58 4 4 Female 40 Control 150 153 87 5 5 Female 50 Case 153 147 53 6 6 Female 50 Control 168 163 62 7 7 Female 60 Case 161 167 34 8 9 10 11 12 13 14 15 16 8 Female 9 Male 10 Male 11 Male 12 Male 13 Male 14 Male 15 Male 16 Male 60 30 30 40 40 50 50 60 60 Control Case Control Case Control Case Control Case Control 180 135 160 152 169 163 182 179 178 178 138 164 155 164 165 183 179 184 53 88 109 76 93 61 73 49 61 biomarker.data= read.csv("biomarkers.csv", header=TRUE) When you have your data in a data frame, you can use the following syntax. The text "Biomarker.1 ~ Disease" is called a formula, and it tells R to perform a t-test with Biomarker.1 as the quantitative response (y) variable, and Disease as the categorical (x) variable. t.test(Biomarker.1 ~ Disease, data=biomarker.data, var.equal=TRUE) Two Sample t-test data: Biomarker.1 by Disease t = -1.8493, df = 14, p-value = 0.08563 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: -30.506601 2.256601 sample estimates: mean in group Case mean in group Control 151.875 166.000 Conclusion: We do not reject the null hypothesis. Biomarker.1 was not significantly different between the Disease groups (p = 0.08563). The 95% confidence interval for the true value of the difference of the means ranges from -30.5 to 2.26. Because the 95% confidence interval includes zero, we do not reject the null hypothesis that the difference between the two groups is zero. Do bears lose weight between winter and spring? Here is another example of a two-sample t-test. We measure the weight of one group of bears in winter, and measure the weight of a different group of bears the following spring. bears.winter=c(300,470,550,650,750,760,800,985,1100,1200) bears.spring=c(280,420,500,620,690,710,790,935,1050,1110) t.test(bears.spring, bears.winter, var.equal=TRUE) > t.test(bears.spring, bears.winter, var.equal=TRUE) Two Sample t-test data: bears.spring and bears.winter t = -0.3732, df = 18, p-value = 0.7134 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: -304.9741 212.9741 sample estimates: mean of x mean of y 710.5 756.5 The p-value=0.7134 is not significant, so we do not reject the null hypothesis that the mean weight of bears in winter is the same as the mean weight of bears in spring. The 95% confidence interval for the true value of the mean is -304.9741 to 212.9741. Because the 95% confidence interval includes zero, we do not reject the null hypothesis that the difference between the two groups is zero. A better design for this study would be to measure the same bears in winter and in spring, to remove the effect of variability among bears from the experiment. We'll do that shortly using a paired t-test. T-test for two independent samples: unequal variance In some cases, we may know or expect that two groups have unequal variance. For example, we may want to compare the mean yield of two processes, where we know that one process is more variable than the other. It may be that a group receiving an active treatment (drug) is more variable in its response than the control (placebo) group. In these cases, we may use an alternative version of the t-test, called Welch's test, which does not assume equal variance of the two treatment groups. Furness and Bryant (1996) compared the metabolic rates of male and female breeding northern fulmars (data described in Logan (2010) and Quinn (2002)). setwd("C:/Users/Walker/Desktop/UCSD Biom 285/Data") furness= read.csv("furness.csv", header=TRUE) furness 1 2 3 4 5 6 SEX Male Female Male Male Male Female METRATE 2950.0 1956.1 2308.7 2135.6 1945.6 1490.5 BODYMASS 875 635 765 780 790 635 7 8 9 10 11 12 13 14 Female Female Female Male Female Male Male Male 1361.3 1086.5 1091.0 1195.5 727.7 843.3 525.8 605.7 668 640 645 788 635 855 860 1005 boxplot(METRATE~SEX, data=furness) The boxplots indicate that the variances of males and females are not equal. So we use the t-test assuming unequal variances. t.test(METRATE~SEX, data=furness, var.equal=F) Welch Two Sample t-test data: METRATE by SEX t = -0.7732, df = 10.468, p-value = 0.4565 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: -1075.3208 518.8042 sample estimates: mean in group Female mean in group Male 1285.517 1563.775 The p-value=0.4565 is not significant, so we do not reject the null hypothesis. Paired t-test for matched samples: Here we'll test the hypothesis that bears lose weight during hibernation. First, we'll assume that different bears are weighed in November and March, and test if their mean weight is different using an ordinary t-test. Then, we'll assume that the same bears are weighed in November and March, and test if their mean weight is different using a paired t-test. The big advantage of measuring the same bears (and using the paired t-test) is that, because each bear serves as its own control, we control for the variability among bears. This has the effect of removing the (unexplained) variability due to variation in weight among the bears. bears.winter=c(300,470,550,650,750,760,800,985,1100,1200) bears.spring=c(280,420,500,620,690,710,790,935,1050,1110) diff= bears.spring - bears.winter stripchart(diff, method="stack") t.test(bears.winter, bears.spring, var.equal=TRUE, paired=TRUE) # In R, we use the argument "paired=TRUE" to do the paired t.test. > t.test(bears.winter, bears.spring, var.equal=TRUE, paired=TRUE) Paired t-test data: bears.winter and bears.spring t = 6.5492, df = 9, p-value = 0.0001053 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: 30.11113 61.88887 sample estimates: mean of the differences 46 The p-value=0.0001053 is significant, so we reject the null hypothesis that the change in weight between winter and spring is zero. One-sample t-test A one-sample t-test tests the hypothesis that the mean of a sample is different from a specified mean. Suppose that we have recruited several students for a study to see if body mass index (BMI) is changed by exercise. In previous studies, the mean BMI of students at baseline (before the exercise) was 18.2. Is the mean BMI =18.2 for the new students at baseline? bmi.new=c(17,17, 18,18,18, 19, 20,20, 21, 22) mean(bmi.new) sd(bmi.new) > mean(bmi.new) [1] 19 > sd(bmi.new) [1] 1.699673 Our null hypothesis is that the bmi of the new students is 18.2. Here's the t-test. t.test(bmi.new, mu=18.2) > t.test(bmi.new, mu=18.2) One Sample t-test data: bmi.new t = 1.4884, df = 9, p-value = 0.1708 alternative hypothesis: true mean is not equal to 18.2 95 percent confidence interval: 17.78413 20.21587 sample estimates: mean of x 19 The p-value=0.17 is not significant, so we do not reject the null hypothesis that the bmi of the new students is 18.2. A little while later we recruit another group of students. Is their mean BMI=18.2? bmi.2=c(18,18,18, 19, 19, 20,20, 21, 22, 22, 23) mean(bmi.2) sd(bmi.2) > mean(bmi.2) [1] 20 > sd(bmi.2) [1] 1.788854 t.test(bmi.2, mu=18.2) > t.test(bmi.2, mu=18.2) One Sample t-test data: bmi.2 t = 3.3373, df = 10, p-value = 0.007525 alternative hypothesis: true mean is not equal to 18.2 95 percent confidence interval: 18.79823 21.20177 sample estimates: mean of x 20 The p-value=0. 007525 is significant, so we reject the null hypothesis that the bmi of this group of students is 18.2. The 95% confidence interval for the true value of the mean is 18.79823 to 21.20177. By default, R assumes that mu=0 unless you specify otherwise. The paired t-test is equivalent to a one sample t-test using the difference between before and after values. Recall that by default, R assumes that mu=0 in a one-sample t-test unless you specify otherwise. # Compare the paired t-test to one sample t-test: diff= bears.spring - bears.winter t.test(diff) > t.test(diff) One Sample t-test data: diff t = -6.5492, df = 9, p-value = 0.0001053 alternative hypothesis: true mean is not equal to 0 95 percent confidence interval: -61.88887 -30.11113 sample estimates: mean of x -46 Log transform If you have outliers and/or non-normal distributions, you may be able to apply a transform to the data to make the distribution more normal, and to reduce the influence of the outliers. Do colon cancer patients have elevated level of the mucin protein in their blood? We measure the level of the protein mucin in the blood of patients with colon cancer and in healthy controls. colon.cancer=c(83, 89, 90, 93, 98) controls=c(99, 100, 103, 104, 141) boxplot(colon.cancer, controls) Let's do a t-test. t.test(colon.cancer, controls, var.equal=TRUE) > t.test(colon.cancer, controls, var.equal=TRUE) Two Sample t-test data: colon.cancer and controls t = -2.258, df = 8, p-value = 0.05389 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: -37.9994752 0.3994752 sample estimates: mean of x mean of y 90.6 109.4 Even though the boxplot shows a clear separation of the two groups, the p-value for the ttest is not significant (p=0.054), due to the outlier (and the small sample size). We could try a log transform of the data to reduce the effect of the outlier. > t.test(log(colon.cancer), log(controls), var.equal=TRUE) Two Sample t-test data: log(colon.cancer) and log(controls) t = -2.5166, df = 8, p-value = 0.036 alternative hypothesis: true difference in means is not equal to 0 95 percent confidence interval: -0.34620596 -0.01512052 sample estimates: mean of x mean of y 4.504971 4.685634 In this case, the log transform yields a p-value of p=0.036, so we reject the null hypothesis and conclude that colon cancer patients have mucin levels different from those of healthy controls.