hw7writeup
advertisement

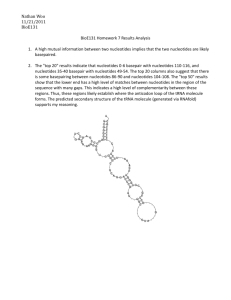
1. Given two indices i and j, mutual information measures the amount of information that I (or the probability distribution of which nucleotides can appear in column i) conveys about the probability distribution of nucleotides in position j. If the mutual information between these two distributions is high, this implies that a lot of information about the probability distribution of column j can be gained by studying the probability of distribution of nucleotides of column i (or vice versa). This would imply that the distributions are not independent, and that the data from those two columns can likely be compressed. 2. Given tRNA’s folded structure, we would expect that there would be some high values of mutual information among certain columns. The columns that have a high mutual information content are those that are more likely to be bonded to one another in the tRNA molecule. So it is likely that the nucleotides in the top 20 columns are bonded. For the nucleotides in the top 50 columns, the probability that they are bonded is somewhat less high, perhaps indicating the presence of an evolutionary change or other type of mutation. As you can see in the attached graphs, when plotting I vs. J, with the size of the plot marks proportional to their information content, the result looks similar to that produced by the probability plots that we used in homework 4. It stands to reason that similar conclusions can be drawn from the plot of mutual information.