Supplemental material: Detailed field studies: We address natural
advertisement

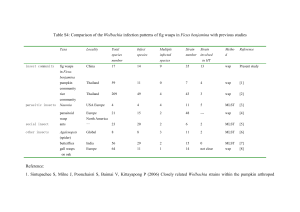
SUPPLEMENTAL MATERIAL: 1. DETAILED FIELD STUDIES: We address natural intrapopulational variation over time in this species through a consideration of two populations: Navafría (NAV) (Sistema Central, Spain), a genetically and geographically isolated pure Spanish population of Cpe, and Sallent de Gállego (SAL) (Pyrenees, Spain), a population inside the HZ (Fig. S1). We have selected these two populations because of their different environmental conditions, and because both are well established populations in which the incidence of Wolbachia has been extensively studied. 1.1. Field Collections Three random samples of the endemism Chorthippus parallelus erythropus were collected from the Iberian population of Navafría (NAV; 42°45'53.31"N;0°20'25.34"W, 1710 m; n = 283) in July, August and September 2005, to analyze the variation in infection frequency in early-, mid- and late-summer populations. These organisms have an annual life cycle, with one generation disappearing by the end of the summer and the next appearing by the end of June of the following year. Similar three-stage sampling was carried out in the hybrid Pyrenean population of Sallent de Gállego (SAL; 42°45'53.31"N;0°20'25.34"W, 1343 m; n = 319) (Fig. 1 and table I). Gonads were dissected and fixed in 100% ethanol. 1.2. DNA Extraction DNA was extracted from whole fixed ovaries and testicles. Each sample was individually dried and immediately frozen in liquid nitrogen before crushing in a mill (Retsch, Germany). Each sample was homogenized in 600 µl of TNES buffer (50 mM Tris-HCl pH 8.0; 400 mM NaCl; 20 mM EDTA pH 8.0; 0.5% SDS) containing proteinase K (0.03% p/v) and incubated at 37ºC overnight. Phenol/chloroform extraction was carried out, followed by ethanol precipitation. Samples were dissolved in Tris-EDTA (tris-HCl 1 mM pH 8.0; EDTA 1 mM pH 8.0) and incubated with RNase A (5 μg/ml) for 2 hours. The final DNA concentration was 50 ng/µl (Martínez et al., 2009; Zabal-Aguirre et al., 2010) 1.3. Wolbachia Detection and Sequencing Wolbachia was detected by PCR amplification of a Wolbachia 16S rRNA gene, using Wolbachia-specific primers (Zabal-Aguirre et al., 2010), followed by a second, nested PCR amplification using strain-specific primers (Martínez-Rodríguez et al., 2013). We performed this highly sensitive technique to limit false-negative data because of the low bacterial densities present during some host ontogenetic stages (Arthofer et al., 2009). PCR reactions were performed in 50 µl containing 2 mM of MgCl2, 0.2 mM of dNTP, 30 pmoles of each primer, 1.25 U of Taq Biotools DNA polymerase, and 4 µl of DNA solution at 50 ng/µl. Thermal cycling conditions were: 30 s at 95°C, followed by 35 cycles of 30 s at 95°C, 1 min at 54°C (for the generalist primers) or 68°C (for the strain-specific primers) and 1 min at 72°C (Zabal-Aguirre et al., 2010; MartínezRodríguez et al., 2013). After these cycles there was a final elongation stage of 10 min at 72°C. 10 µl of each amplification product were electrophoretically separated on 0.7% agarose gel. Gels were stained with ethidium bromide. To confirm the strain characterization, the Multilocus Sequence Typing (MLST) and wsp (Wolbachia surface protein) gene characterization systems were used. GatB, coxA, hcpA, ftsZ, fbpA and wsp genes were amplified as previously described (Baldo et al., 2006), with modifications: PCR reactions were performed in 50 µl containing 2 mM of MgCl2, 0.2 mM of dNTP, 30 pmoles of each primer, 1.25 U of Taq BIOTAQ™ DNA polymerase (Bioline) and 4 µl of DNA solution (50 ng/µl). Amplified genes were purified by Illustra GFX PCR DNA and Gel Band Purification Kit (GE Healthcare) or the ExoSAP-IT method (GE Healthcare). Sequencing was performed by Stabvida (www.stabvida.com). The 16S rRNA, MLST, and wsp sequences generated in this study have been deposited in the GenBank database under accession numbers JN698878- JN698882 and JN701984- JN701990. Bayesian likelihood was inferred using a Markov Chain-Monte Carlo variant run under the MrBayes 3.2.1 program (Ronquist & Huelsenbeck, 2003). Phylogenies based on single and concatenated MLST genes and wsp were reconstructed. jModeltest (Posada, 2008) was used to distinguish the appropriate model of evolution, the best likelihood score being evaluated by AIC criteria (Akaike, 1974). The model selected were GTR+I+G (General time reversible model, including Gamma and Proportion Invariant corrections) for concatenated MLST, ftsZ, gatB, and wsp; GTR+G for coxA and hcpA, and HKY+I+G (Hasegawa, Kishino and Yano model, including Gamma and Proportion Invariant corrections) for fbpA. Bayesian analysis was carried out with 106 generations and a sample frequency of 100. The first 5,000 trees were discarded as burn-in. 1.4. Statistical Analysis and Temperature Data Data were analyzed with SPSS Statistics 19.0. Wilson’s correction was used to estimate the infection proportions and their corresponding SEs (Wilson, 1927). Proportions of crosstabulated variable categories were compared using the χ2 test (table II). Temperatures at NAV were provided by the INM (Spanish National Meteorological Institute), and those from SAL were kindly provided by José A. Villacampa from ARAMON S.L. (Formigal, Spain). Mean temperatures were compared with a one-way ANOVA. The mean temperatures for July and August were different for NAV and SAL, whilst those for September were similar (fig. 8). NAV temperatures frequently exceeded 30ºC during July and August, occasionally rising above 35ºC. Temperatures in SAL were significantly lower than in NAV, except September (ANOVA: Global data: F = 55.23; d.f. = 1, 152; p ≈ 0.000. July: F = 26.608; d.f. = 1, 60; p ≈ 0.000. August: F = 62.840; d.f. = 1, 60; p ≈ 0.000. September: F = 0.496; d.f. = 1, 28; p = 0.487), varying between 20°C and 25°C. (Fig. S4). Infection proportions did not change over the summer (see above). SUPPLEMENTAL FIGURES : Figure S1: Populations of Chorthippus parallelus: Navafría (NAV) and Sallent de Gállego (SAL). Figure S2: Rooted Bayesian tree of the Wolbachia 16s rRNA sequences analyzed in this study. Notice that Wolbachia strains (grey) belong to the F and B supergroups. Figure S3: Unrooted Bayesian trees of the Wolbachia MLST genes and Wsp gene analyzed in this study. Notice that Wolbachia strains (grey) belong to the F and B supergroups. Figure S4: Temperatures in the Chorthippus parallelus population locations: Navafría (NAV, black) and Sallent de Gállego (SAL, grey). Progression of single infection in C. parallelus. Notice that infected and uninfected proportions are complementary. a) Change in proportions of uninfected males after 4000 generations (stable equilibrium) as a function of the initial uninfected male frequency (Pm0) and maternal transmission (where µ = 0 represents perfect transmission). The initial female frequency was P f0 = 0.5. b) Change in proportions of uninfected females as a function of initial uninfected female (Pf0) frequency and maternal transmission. Initial male frequency was Pm0 = 0.5. c) and d) Dependence of threshold value on H (where H = 0 represents total incompatibility) and µ (where µ = 0 represents perfect transmission). Initial male and female proportions were Pm0 = 0.5 and Pf0 = 0.5. Note the discontinuity in the predicted infection proportions for parameter values. This discontinuity appears irrespective of the initial infection proportions. Figure S5: SUPPLEMENTAL TABLES Population Latitude Longitude Altitude (m) n Inf F Inf B Inf FB Uninfected NAV 40° 59´02´´N 3° 49´00´´ W 1710 283 0.64 ± 0.06 0.10 ± 0.04 0.22 ± 0.05 0.04 ± 0.02 SAL 42° 45´57´´ N 0° 20´ 33´´ W 1343 321 0.02 ± 0.02 0.23 ± 0.05 0.73 ± 0.05 0.02 ± 0.02 Table S-1: Wolbachia infection proportions ± 2 SE in Navafría (NAV) and Sallent de Gállego (SAL) populations of Chorthippus parallelus, independent of sex and month of sampling. Let n be the number of individuals. Wilson’s correction was used to estimate the infection proportions and the corresponding SEs (Wilson, 1927). Sex Infection Kind All individuals Males Females July August Strain Proportion B F BF 0.34 ± 0.10 0.99 ± 0.04 0.02 ± 0.04 92 0.63 ± 0.10 Uninfected 0 B Infected F BF 0.01 ± 0.04 0.01 ± 0.04 0.45 ± 0.15 0.98 ± 0.07 0.00 ± 0.06 Infected Uninfected 0 B Infected F BF Uninfected 0 n 0.53 ± 0.15 0.02 ± 0.07 0.02 ± 0.07 0.22 ± 0.13 1.00 ± 0.06 0.04 ± 0.09 0.73 ± 0.14 0.00 ± 0.06 0.00 ± 0.06 47 45 September Proportion 0.99 ± 0.03 n 0.01 ± 0.03 0.01 ± 0.03 0.23 ± 0.12 1.00 ± 0.05 0.02 ± 0.06 0.75 ± 0.12 0.00 ± 0.05 0.00 ± 0.05 0.23 ± 0.11 0.99 ± 0.05 0.00 ± 0.04 0.75 ± 0.11 0.01 ± 0.05 Proportion n 0.23 ± 0.08 0.15 ± 0.07 0.99 ± 0.03 0.01 ± 0.03 0.02 ± 0.04 122 107 0.75 ± 0.08 0.82 ± 0.08 0.01 ± 0.05 0.01 ± 0.03 0.02 ± 0.04 0.20 ± 0.12 0.98 ± 0.07 0.04 ± 0.08 57 49 0.75 ± 0.13 65 0.02 ± 0.07 0.02 ± 0.07 0.10 ± 0.08 1.00 ± 0.05 0.00 ± 0.05 0.90 ± 0.09 58 0.00 ± 0.05 0.00 ± 0.05 Table S-2: Wolbachia infection proportions ± 2 SE in the Sallent de Gállego (SAL) population of Chorthippus parallelus. Let n be the number of individuals. Wilson’s correction was used to estimate the infection proportions and the corresponding SEs (Wilson, 1927). Sex Infection Kind July Strain B All individuals Infected Uninfected Males Females Infected F BF 0 B F BF Uninfected 0 B Infected F BF Uninfected 0 August Proportion n 0.06 ± 0.06 1.00 ± 0.06* 0.57 ± 0.10 0.40 ± 0.10* 0.00 ± 0.06* 0.00 ± 0.03* 0.02 ± 0.07 1.00 ± 0.05* 0.58 ± 0.14* 0.4 ± 0.14* 0.00 ± 0.05* 0.00 ± 0.05* 0.10 ± 0.10 1.00 ± 0.05 0.56 ± 0.14 0.33 ± 0.14 0.00 ± 0.05 0.00 ± 0.05 September Proportion n 0.11 ± 0.07 98 50 48 0.98 ± 0.04* 0.68 ± 0.10 0.19 ± 0.08* 0.02 ± 0.04* 0.02 ± 0.04* 0.08 ± 0.09 0.98 ± 0.07* 0.79 ± 0.12 0.11 ± 0.10 0.02 ± 0.07* 0.02 ± 0.07 0.14 ± 0.10 0.98 ± 0.07 0.58 ± 0.14 0.26 ± 0.12 0.02 ± 0.07 0.02 ± 0.04 Proportion n 0.13 ± 0.08 0.91 ± 0.07* 0.68 ± 0.10 88 0.10 ± 0.07* 0.09 ± 0.07* 0.09 ± 0.07* 0.07 ± 0.10 0.85 ± 0.13* 0.73 ± 0.14* 47 41 0.05 ± 0.09* 97 50 0.15 ± 0.13* 0.15 ± 0.12* 0.17 ± 0.12 0.96 ± 0.08 0.63 ± 0.14 0.15 ± 0.11 47 0.04 ± 0.08 0.04 ± 0.08 Table S-3: Wolbachia infection proportions ± 2 SE in the Navafría (NAV) population of Chorthippus parallelus erythropus. Asterisks denote significant differences between infection levels through the summer. Let n be the number of individuals. Wilson’s correction was used to estimate the infection proportions and the corresponding SEs (Wilson, 1927). SUPPLEMENTAL REFERENCES: Akaike, H., (1974). A new look at the statistical model identification. IEEE Trans. Aut. Control 19, 716–723. Arthofer, W., Riegler, M., Schneider, D., Krammer, M., Miller, W.J., Stauffer. C. (2009) Hidden Wolbachia diversity in field populations of the European cherry fruit fly, Rhagoletis cerasi (Diptera, Tephritidae). Mol. Ecol. 18,: 3816-3830. doi:10.1111/j.1365-294X.2009.04321.x Baldo, L., Hotopp, J. C. D., Jolley, K. A., Bordenstein, S. R., Biber, S. A., Choudhury, R. R., Hayashi, C., Maiden, M. C. J., Tettelin, H., Werren, J. H. (2006). Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Applied and Environmental Microbiology 72, 7098-7110. Martinez-Rodriguez, P., Hernandez-Perez, M., Bella, J.L. (2013) Detection of Spiroplasma and Wolbachia in the bacterial gonad community of Chorthippus parallelus. Microbial Ecology 16, 16. Martínez, P., Del Castillo, P., Bella, J. L., (2009). Cytological detection of Wolbachia in squashed and paraffin embedded insect tissues. Biotechnic & Histochemistry 84, 347-353. Posada, D., (2008). jModelTest: Phylogenetic model averaging. Molecular Biology and Evolution 25, 1253-1256, doi:10.1093/molbev/msn083 Ronquist, F., Huelsenbeck, J. P., (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572-1574, doi:10.1093/bioinformatics/btg180. Wilson, E., (1927). Probable inference, the law of succession, and statistical inference. Journal of the American Statistical Association 22, 209-212 Zabal-Aguirre, M., Arroyo, F., Bella, J. L., (2010). Distribution of Wolbachia infection in Chorthippus parallelus populations within and beyond a Pyrenean hybrid zone. Heredity 104, 174-184, doi:10.1038/hdy.2009.106.