Supplementary Material - Journal of the American College of
advertisement

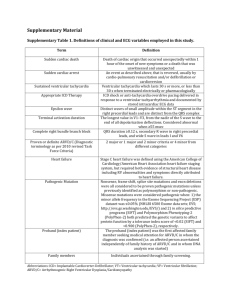
Supplementary Material Supplementary Methods Study design. Four-to-six-week-old wild-type C57BL6/J mice were injected with 3.5x1010 viral genomes (vg) encoding Luc, wild-type human PKP2, or the C-terminal deletion mutant version R735X. Animals were divided into two groups: group A = trained, and group B = sedentary. Training in group A started 2 weeks after injection and continued for 8 consecutive weeks. At the end of that period, Luc, PKP2 and R735X mice were imaged by Cardiac Magnetic Resonance (CMR) and euthanized for heart sampling. Animals in group B were analysed by CMR 6 and 10 months after infection with AAV particles. Animals and endurance training protocol. All animals were maintained and handled according to the recommendations of the CNIC Institutional Ethics Committee. See Supplemental Methods for animal procedures. Wild-type 4-5-week-old male C57BL/6J mice were obtained from Charles River Laboratories. Mice were individually housed in wire-bottomed cages in a temperaturecontrolled room (22±0.8°C) with a 12 h light–dark cycle and a relative humidity of 55±10%. The mice had free access to food and water. All mice were adapted to water before beginning the eight-week endurance swimming training program. Adaptation consisted of keeping the animals in shallow water at 30 to 32ºC, with the purpose of reducing the environmental stress without promoting any physical training adaptations. Endurance training took place during the dark phase of the light–dark cycle to coincide with the period of animals’ maximum activity, and training intensity was increased gradually. Mice swam in water at a depth of 25 cm in a glass container (60×30×45 cm) at 30 to 32ºC. The animals were progressively familiarized with swimming during the first 2 weeks by increasing the swimming time by 5 min per day, reaching 45 min per day at the end of the second week. Daily swimming duration was then increased by 15 min each week, and held at 90 min per day during the final 3 weeks. Mice were allowed to swim at their own pace, and the water was gently bubbled to ensure that mice swam rather than floating. Week 1 Week 2 Week 3 Week 4 Week 5 Week 6 Week 7 Week 8 Time in minutes 5 10 15 20 25 0 0 25 30 35 40 45 0 0 45 45 45 45 45 0 0 60 60 60 60 60 0 0 75 75 75 75 75 0 0 90 90 90 90 90 0 0 90 90 90 90 90 0 0 90 90 90 90 90 End Random analysis of distance swum revealed a Gaussian distribution with a coefficient of variation of 8.4% (Supplementary Figure 6), confirming homogeneous training intensity across groups. CMR imaging. During CMR evaluation, animals were anaesthetized with isoflurane and were monitored for core body temperature, cardiac rhythm and respiration rate using a CMR compatible monitoring system. In vivo cardiac images were acquired using an Agilent VNMRS DD1 7T MRI system (Santa Clara, California, USA). A k-space segmented ECG-triggered cine gradient-echo sequence was used. After shimming optimization, cardiac four-chamber and left two-chamber views were acquired and used to plan the short axis sequence. Mice were imaged with the following parameter settings: number of slices, 13; slice thickness, 0.8 mm; gap, 0 mm; matrix size, 256x256; field of view, 30x30 mm2; gating: ECG and respiratory triggered; cardiac phases, 20; averages, 4; effectiveTE, ~1.8 ms, minimumTR, 7 ms; flip angle, 25º; trigger delay, 2 ms; trigger window, 8 ms; dummy scans, 2. CMR data analysis. All CMR images were analysed using dedicated software (QMass MR v.7.5, Medis, Leiden, the Netherlands). Images were analysed by two experienced observers blinded to the study allocation. All CMR images were of good quality and could be analysed. The short-axis data set was analysed quantitatively by manual detection of endocardial borders in end-diastole and end-systole, with exclusion of papillary muscles and trabeculae, in order to obtain both left and right end-diastolic volume, end-systolic volume and ejection fraction. The right ventricle was assessed qualitatively by evaluating the wall motion in the cine modus in short- and long-axis views. The right ventricle was divided into 11 segments(1,2). Wall motion was classified as abnormal in the presence of akinesia, dyskinesia (in ventricular systole), or bulging (in ventricular diastole). Surface ECG. Mice were anaesthetized using isoflurane inhalation (0.8-1.0% volume in oxygen), and efficacy of the anaesthesia was monitored by watching breathing speed. Four-lead surface ECGs were recorded, for a period of 5 minutes, from subcutaneous 23-gauge needle electrodes attached to each limb using the MP36R amplificator unit (BIOPAC Systems). During offline analysis, lead II was used for QRS duration using AcqKnowledge 4.1 analysis software. A representative 30s segment of the recording was averaged to obtain the signal-averaged ECG. Then, QRS duration was measured as the time interval between the earliest moment of deviation from baseline and the moment when the Swave returned to the isoelectric line. QRS duration from endurance-trained R735X-infected mice was compared to those of contemporaneous animals infected with wild type PKP2 and Luc pooled together. Similarly, QRS duration from sedentary AAV-R735X mice at 10 months (long-term) was compared to those of the 2 groups of controls pooled together. AAV vector production and purification. AAV vectors were all produced by the triple transfection method, using HEK 293A cells as described previously(3). AAV plasmids were cloned and propagated in the Stbl3 E. coli strain (Life Technologies). Shuttle plasmids pAAV-PKP2, pAAV-R735X and pAAV-Luc were derived from pAcTnT (a gift from Dr B.A. French) and packaged into AAV-9 capsids with the use of helper plasmids pAdDF6 (providing the three adenoviral helper genes) and pAAV2/9 (providing rep and cap viral genes), obtained from PennVector. Shuttle vectors were generated by direct cloning (GeneScript) of synthesized fragments from NheI-SalI into pAcTnT cut with the same restriction enzymes. The AAV shuttle and helper plasmids were transfected into HEK 293A cells by calcium-phosphate co-precipitation. A total of 840µg plasmid DNA (mixed in an equimolar ratio) was used per Hyperflask (Corning) seeded with 1.2x108 cells the day before. Seventy-two hours after transfection, the cells were collected by centrifugation and the cell pellet was resuspended in TMS (50 mM Tris HCl, 150 mM NaCl, 2 mM Cl2Mg) on ice before digestion with DNase I and RNaseA (0.1 mg/mL each; Roche) at 37 °C for 60 minutes. Clarified supernatant containing the viral particles was obtained by iodixanol gradient centrifugation(4). Gradient fractions containing virus were concentrated using Amicon UltraCel columns (Millipore) and stored at -70ºC. Determination of AAV vector titer. Titers for the AAV vectors (vg per ml) were determined by quantitative real-time PCR as described(5). See primers used below. Known copy numbers (105–108) of the respective plasmid (pAAV-PKP2, pAAVR735X and pAAV-Luc) carrying the appropriate complementary DNA were used to construct standard curves. Quantitative real-time PCR. For mouse tissue analysis, total RNA was isolated from mouse hearts and liver using the Direct-zol RNA Miniprep Kit (Zymo) and reverse transcribed with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Complementary DNAs were analyzed by real time PCR using the Power SYBR® Green PCR Master Mix (Applied Biosystems). Amplification, detection and data analysis were completed with the ABI PRISM® 7900HT Sequence Detection System. The crossing threshold values for individual mRNAs were normalized to GADPH. Changes in the expression of mRNA were expressed as the fold change relative to the control. We used the following primers: PKP2 F, 5′ AGAGCAGCAGCACAGTACAG3′; PKP2 R, 5’ CGCCAGGCATACTGGTATTC3′; GapdhF, 5′ TTGATGGCAACAATCTCCAC 3′; GapdhR, 5′ CGTCCCGTAGACAAAATGGT 3′; Pkp2 F, 5’ CAGAACAGGGCTTCCAGGTC 3’; Pkp2R, 5’ CTGCTCGCTCCAGAGTCATC3’; eGFPF,5’ AGCTGGACGGCGACGTAAAC 3’; eGFPR 5’ AAGATGGTGCGCTCCTGGAC 3’ Immunofluorescence At the end of exercise training, hearts were collected and fixed in 2% paraformaldehyde in phosphate-buffered saline (PBS) for 1 hour at 4°C. After washing three times in PBS for 5 min, hearts were equilibrated with 15% sucrose in PBS for 1 hour and incubated in 30% sucrose overnight at 4°C. Samples were included in OCT (Tissue-tek OCT compound, Sakura-4583), and 5μm cryosections were prepared for immunofluorescence. Tissues were permeabilized with PBS-0.2% Triton (Sigma, T9284) for 10 min, and washed in PBS-0.1% Triton (PBST). Tissues were blocked with 10% goat serum in PBST for 1 hour. Primary antibodies against plakoglobin (610253, BD) and PKP2 (651167, Progen) required antigen retrieval. Antibodies against sarcomeric cardiac α-actinin (A7732, Sigma), Cx43 (C6219, Sigma), and eGFP (632592, Clonthech) were diluted in PBST, and slides were incubated with antibodies overnight at 4°C. After washes, slides were incubated with Cy3-conjugated or Alexa Fluor 488-conjugated goat anti-mouse secondary antibodies for 45 min in the presence of DAPI. Vectashield mounting medium without DAPI (Vectors Lab.; H-1000) was applied to all slides. Fluorescence images were obtained with a Nikon A1-R inverted confocal microscope. The percentage of positive cells in the total tissue area was calculated using ImageJ after segmentation of the cells and measurement of signal intensities. Transmission electron microscopy Immediately after excision, tissues were fixed in 4% formahaldehyde: 1% glutaraldehyde in cacodylate buffer, and postfixed in 1% osmium tetroxide. Tissues were then washed in PBS, dehydrated through graded alcohols followed by acetone, and then infiltrated with Durcupan ACM Fluka resin and polymerized at 60 ºC for 48h. Blocks were cut with a Leica ultracut UCT ultramicrotome (Leica, Heerbrugg, Switzerland) and sections (60–70 nm) were mounted onto 200-mesh grids. Sections were stained with a 2% solution of aqueous uranyl acetate for 10 min, followed by lead citrate staining for 10 min. Stained sections were viewed with a JEOL JEM-1010 transmission electron microscope (Tokyo, Japan) operating at 100 kV. Images were acquired with a GATAN (Orius 200SC) digital camera. Statistics. Experiments were designed to use the minimum number of mice needed to give sufficient statistical power, and the numbers of animals used are comparable to published literature for the same assays(6). No animals were excluded from the analyses. Data were analysed by one-way ANOVA, two-way ANOVA and Student’s t test. Relative risk analysis was assessed by two-tailed Fisher’s exact test. Reliability of CMR measures was assessed by inter-observer intraclass correlation coefficient (absolute agreement) and mean bias. Error bars represent SEM. In all corresponding figures, ∗ p< 0.05, ∗∗ p< 0.01, and ∗∗∗ p< 0.001, and ns p> 0.05. Supplementary Video 1. CMR ventricular short axis (SAX) slices covering from apex to base in a control mouse expressing wild-type PKP2. Supplementary Video 2. CMR ventricular short axis (SAX) slices covering from apex to base in an R735X mutant mouse. Note the dyskinesis in the basal portion of the right ventricle (RV). Supplementary Video 3. CMR ventricular short axis (SAX) slices covering from apex to base in an R735X mutant mouse. Note the severe dyskinesis in the right and left ventricle. Supplementary Figure 1. Efficient expression of target genes in heart. qRT-PCR analysis of total mRNA isolated from heart and liver 10 weeks after AAV infection. Elevations of EGFP, PKP2 and R735X mRNA expression levels were evident in hearts relative to the levels in liver. Supplementary Figure 2. Long-term expression of target genes in heart. qRT-PCR analysis of total mRNA isolated from heart and liver 10 months after AAV infection. Elevations of PKP2 and R735X mRNA expression levels were evident in hearts relative to the levels in liver. Supplementary Figure 3. Electrocardiographic findings. QRS complex duration in R735X mice and controls (PKP2 and Luc infected). Panel A shows the results of longterm sedentary mice while panel B corresponds to mice after finishing the endurance training. ms= miliseconds. Supplementary Figure 4. Desmosomal structure and protein components in which mutation is linked to ARVC: Desmoplakin (DSP) ARVC8, Plakophilin-2 (PKP2) ARVC9, Desmoglein-2 (DSG2) ARVC10, Desmocollin-2 (DSC2) ARVC11, and Plakoglobin (JUP) ARVC12. Supplemetary Figure 5. Cx43 mRNA and total protein levels remain constant in AAV transduced hearts. (A) Hearts were lysed after swimming training, and protein extract was probed by western blot for Cx43 and tubulin. Each lane represents a lysate from an individual mouse. (B) Quantitative PCR of hearts infected with AAV expressing Luciferase, PKP2 and R735X. Supplementary Figure 6. Uniformity of training intensity. Analysis of distance swum, calculated from randomly taken time-lapse images, including 1000 consecutive frames. The data fit a Gaussian distribution (passed DÁgostino & Pearson omnibus normality test; alpha=0.05). References 1. Sievers B, Addo M, Franken U, Trappe HJ. Right ventricular wall motion abnormalities found in healthy subjects by cardiovascular magnetic resonance imaging and characterized with a new segmental model. Journal of cardiovascular magnetic resonance : official journal of the Society for Cardiovascular Magnetic Resonance 2004;6:601-8. 2. Bluemke DA, Krupinski EA, Ovitt T et al. MR Imaging of arrhythmogenic right ventricular cardiomyopathy: morphologic findings and interobserver reliability. Cardiology 2003;99:153-62. 3. Xiao X, Li J, Samulski RJ. Production of high-titer recombinant adenoassociated virus vectors in the absence of helper adenovirus. Journal of virology 1998;72:2224-32. 4. Hauswirth WW, Lewin AS, Zolotukhin S, Muzyczka N. Production and purification of recombinant adeno-associated virus. Methods in enzymology 2000;316:743-61. 5. Prasad KM, Xu Y, Yang Z, Toufektsian MC, Berr SS, French BA. Topoisomerase inhibition accelerates gene expression after adeno-associated virusmediated gene transfer to the mammalian heart. Molecular therapy: the journal of the American Society of Gene Therapy 2007;15:764-71. 6. Kirchhof P, Fabritz L, Zwiener M et al. Age- and training-dependent development of arrhythmogenic right ventricular cardiomyopathy in heterozygous plakoglobin-deficient mice. Circulation 2006;114:1799-806.

![Historical_politcal_background_(intro)[1]](http://s2.studylib.net/store/data/005222460_1-479b8dcb7799e13bea2e28f4fa4bf82a-300x300.png)