molecular interactions between necrotrophic leaf blotch
advertisement
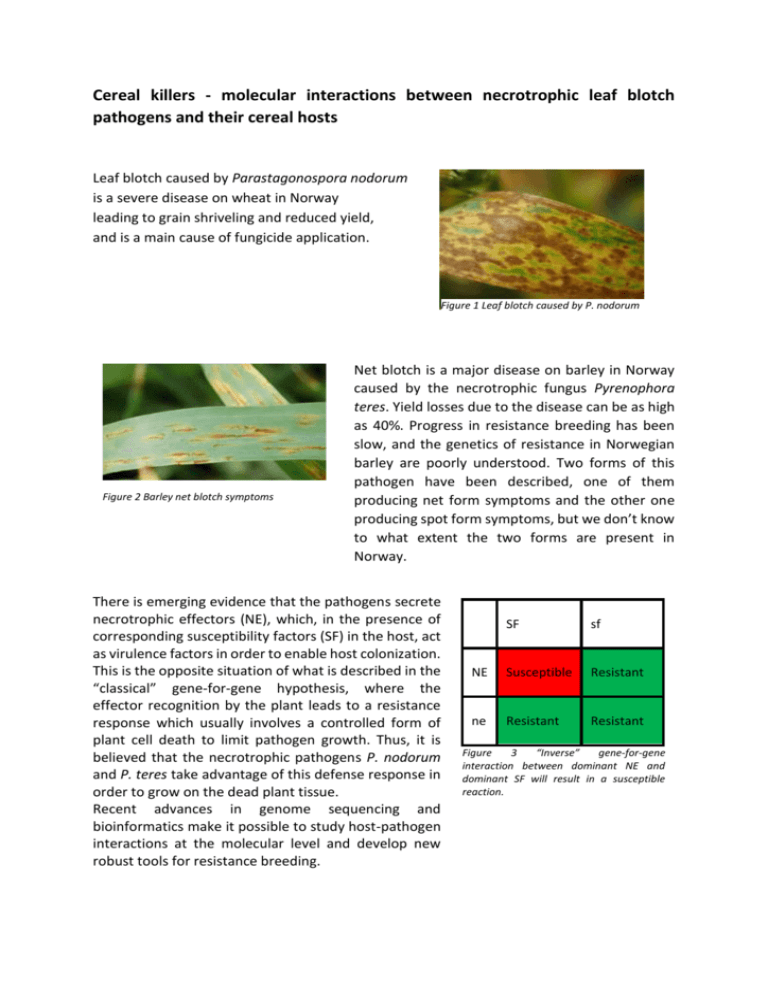
Cereal killers - molecular interactions between necrotrophic leaf blotch pathogens and their cereal hosts Leaf blotch caused by Parastagonospora nodorum is a severe disease on wheat in Norway leading to grain shriveling and reduced yield, and is a main cause of fungicide application. Figure 1 Leaf blotch caused by P. nodorum Figure 2 Barley net blotch symptoms Net blotch is a major disease on barley in Norway caused by the necrotrophic fungus Pyrenophora teres. Yield losses due to the disease can be as high as 40%. Progress in resistance breeding has been slow, and the genetics of resistance in Norwegian barley are poorly understood. Two forms of this pathogen have been described, one of them producing net form symptoms and the other one producing spot form symptoms, but we don’t know to what extent the two forms are present in Norway. There is emerging evidence that the pathogens secrete necrotrophic effectors (NE), which, in the presence of corresponding susceptibility factors (SF) in the host, act as virulence factors in order to enable host colonization. This is the opposite situation of what is described in the “classical” gene-for-gene hypothesis, where the effector recognition by the plant leads to a resistance response which usually involves a controlled form of plant cell death to limit pathogen growth. Thus, it is believed that the necrotrophic pathogens P. nodorum and P. teres take advantage of this defense response in order to grow on the dead plant tissue. Recent advances in genome sequencing and bioinformatics make it possible to study host-pathogen interactions at the molecular level and develop new robust tools for resistance breeding. SF sf NE Susceptible Resistant ne Resistant Resistant Figure 3 “Inverse” gene-for-gene interaction between dominant NE and dominant SF will result in a susceptible reaction. Focus of the project: We use a combination of classical inoculation and infiltration experiments in the field and in the greenhouse and next-generation sequencing and bioinformatics such as QTL mapping to explore the molecular mechanisms that are decisive for resistance or susceptibility: - QTL mapping of resistance/susceptibility loci in different mapping populations of wheat and barley: o Infection at seedling stage under greenhouse conditions and as adult plants in the field. Assessment of disease severity for each genotype. If resistance of adult plants can be already predicted at the seedling stage, this will be a useful screening tool for breeders. o Construction of genetic maps based on high-density SNP markers o QTL mapping to find genetic regions that are associated with resistance or susceptibility - Identification of necrotrophic effectors and corresponding host susceptibility factors by infiltration bioassays of wheat and barley seedlings - ddRAD (double digest restriction associated DNA tags) genotyping-by-sequencing of 400 P. teres isolates in order to unravel the genetic diversity and structure of the Norwegian P. teres population Suggested master thesis topics: - Seedling inoculations with culture filtrates of selected isolates in order to screen for differential disease reactions and to identify potential NEs. o This method can be used to identify isolates which secrete potential NEs. By growing the fungus in a liquid medium it is possible to collect the secreted proteome. These so-called culture filtrates are then infiltrated into the host and it is observed whether differential disease symptoms are developing. By fractioning the filtrates according to protein size by LC-MS and infiltrating these filtrate fractions into the plant, it is possible to get an idea of the size of the potential NE. Additionally, we can use the phenotypic data from infiltration experiments to identify effector sensitivity loci in the plant by QTL/association mapping. - Sequencing and haplotype investigation of NE genes from a collection of Norwegian P. nodorum isolates. This can tell us something about the variation and also population structure of the pathogen. - Characterization and purification of potential NEs by chromatography and/or ultrafiltration methods - Deep sequencing of selected isolates in search of candidate effector loci - QTL and association mapping for resistance/susceptibility loci in wheat/barley, based on the genetic maps and phenotypic data from field and seedling trials o Extensive seedling and adult plant phenotypic data is already available, but it is possible to screen more isolates on seedlings as well as being involved in data analysis and association mapping. - Association mapping of effector genes in P. teres o A collection of 365 Norwegian and 40 international P. teres isolates is available for resistance screenings on selected barley lines. These isolates are currently being ddRAD genotyped in order to obtain SNP markers, enabling association mapping for candidate effector loci in the pathogen. - Validation of molecular marker-assays on plant breeding materials. Depending on the topic and on the field of interest of the student, the work will involve a combination of greenhouse work, molecular lab work and bioinformatics. The cereal leaf blotch group: Morten Lillemo (researcher) morten.lillemo@nmbu.no Andrea Ficke (researcher; NIBIO) andrea.ficke@nibio.no Anja Ruud (PhD student) anja.ruud@nmbu.no Ronja Wonneberger (PhD student) ronja.wonneberger@nmbu.no