Assay variability - F statistic to compare two variances
advertisement

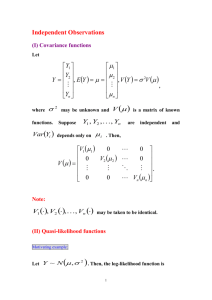
Assay variability: F statistic to compare two variances
One of my clients was having problems with an assay that gave highly variable results when identical
specimens were assayed. They modified the assay to try to reduce the variance. Then they did an
experiment to see if they had been successful in reducing the variance. They took a single, well-mixed
specimen and divided it into 20 test tubes. They randomly split the 20 test tubes into two groups of 10,
and assayed 10 specimens with assay A (the old version) and 10 specimens with assay B (the new
version).
We'll generate some data to simulate their experiment. The R function rnorm(n, mean = 0, sd = 1)
generates n samples from a normal distribution with the mean and standard deviation that you specify.
By default, mean=0 and sd=1, which is a standard normal distribution.
So this code will generate n=10 specimens with mean=20 and sd=2.
rnorm(n=10, mean=20, sd=2)
Let's generate simulated data for the assays. For assayA, we'll have sd=2, and for assayB we’ll have sd=1.
So assayB has half the standard deviation of assayA.
assayA=rnorm(n=10, mean=20, sd=2)
assayA
assayB=rnorm(n=10, mean=20, sd=1)
assayB
sd(assayA)
sd(assayB)
var(assayA)
var(assayB)
var(assayA)/var(assayB)
Here are values I got when I generated the data.
> assayA
[1] 19.97186 14.14566 19.64671 18.42836 18.97717 18.53903 19.45039 22.06924
[9] 17.29014 18.13014
> assayB
[1] 19.64612 19.42008 21.17786 20.87628 22.52887 21.16597 18.58258 18.78492
[9] 19.01121 21.51138
> sd(assayA)
[1] 2.04525
> sd(assayB)
[1] 1.349365
How can we determine if assayB has significantly less variability that assayA? We use the F test to
compare two variances. In R the F test is implemented in the function var.test(). Let's use var.test() on
our assay data.
var.test(assayA, assayB)
> var.test(assayA, assayB)
F test to compare two variances
data: assayA and assayB
F = 2.2974, num df = 9, denom df = 9, p-value = 0.2312
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.5706379 9.2492583
sample estimates:
ratio of variances
2.297385
Oops. That's embarrassing. The p-value for the difference of the two variances is p=0.2313, which is not
significant. Maybe the random samples just didn't differ much? But the test output says that the ratio of
the variance is 2.297, and we saw the difference in the standard deviations:
> sd(assayA)
[1] 2.04525
> sd(assayB)
[1] 1.349365
par(mfrow=c(1,2))
plot(assayA,ylim=c(0,30))
abline(h=mean(assayA))
for (i in 1: length(assayA)){
lines(c(i,i),c(mean(assayA), assayA[i]))}
plot(assayB,ylim=c(0,30))
abline(h=mean(assayB))
for (i in 1: length(assayB)){
lines(c(i,i),c(mean(assayB), assayB[i]))}
par(mfrow=c(1,1))
Let's try generating another 10 observations from each assay and see what happens.
assayA2=rnorm(n=10, mean=20, sd=2)
assayA2
assayB2=rnorm(n=10, mean=20, sd=1)
assayB2
var.test(assayA2, assayB2)
> var.test(assayA2, assayB2)
F test to compare two variances
data: assayA2 and assayB2
F = 3.7586, num df = 9, denom df = 9, p-value = 0.06158
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.933594 15.132279
sample estimates:
ratio of variances
3.758644
The p-value is a little better, at p=0.06, but still not significant, even though we know that the standard
deviation of Assay A is twice that of Assay B. Maybe our sample size is too small. Let's do a simulation to
determine the power to detect a reduction of sd from 2 to 1.
n = 10
pvalue.list = c()
for (i in 1:1000)
{
assayA.sample = rnorm(n, mean=20, sd=2)
assayB.sample = rnorm(n, mean=20, sd=1)
pvalue.list[i] = var.test(assayA.sample, assayB.sample)$p.value
pvalue.list
}
# Plot the pvalue.list
hist(pvalue.list, xlim= c(0, 1), breaks=seq(0,1,.05), ylim=c(0,1000))
# What percent of the 1000 simulated samples give a p-value less than 0.05?
100*sum(sort(pvalue.list)<.05)/length(pvalue.list)
[1] 51.5
So we only detect a significant difference in the variance of assayA versus assayB in 51.5 percent of the
samples. That is, our power is 51.5%. The probability that we will get a significant p-value, even though
there is a two-fold difference in the standard deviation, is only 51.5%.
How many specimens do we need to have to get 80% power? Let's try 20 specimens for each assay,
giving a total of 20+20=40.
n = 20
pvalue.list = c()
for (i in 1:1000)
{
assayA.sample = rnorm(n, mean=20, sd=2)
assayB.sample = rnorm(n, mean=20, sd=1)
pvalue.list[i] = var.test(assayA.sample, assayB.sample)$p.value
pvalue.list
}
# Plot the pvalue.list
hist(pvalue.list, xlim= c(0, 1), breaks=seq(0,1,.05), ylim=c(0,1000))
# What percent of the 1000 simulated samples give a p-value less than 0.05?
100*sum(sort(pvalue.list)<.05)/length(pvalue.list)
[1] 84
That looks better. So to detect a two-fold reduction in the assay standard deviation with 84% power, we
need 20 specimens per assay.
Now let's go back to the assay and simulate a single experiment with 20 specimens per assay and test
for a difference in variance.
assayA3=rnorm(n=20, mean=20, sd=2)
assayA3
assayB3=rnorm(n=20, mean=20, sd=1)
assayB3
sd(assayA3)
sd(assayB3)
Here are values I got when I generated the data.
> assayA3=rnorm(n=20, mean=20, sd=2)
> assayA3
[1] 20.23055 22.28764 20.47531 22.38541 21.89745 22.69879 20.26005 18.21365 16.79730 18.70380
[11] 16.67199 20.02466 22.60145 18.22082 20.03363 19.28665 20.80830 19.43308 21.02003 21.69179
> assayB3=rnorm(n=20, mean=20, sd=1)
> assayB3
[1] 18.44107 20.54730 21.32686 19.32063 19.38746 19.99892 20.85614 20.29317 21.89105 19.31109
[11] 19.98739 20.12810 21.60681 21.80682 19.12465 20.79461 18.71171 20.15293 20.27934 20.09736
>
> sd(assayA3)
[1] 1.820626
> sd(assayB3)
[1] 0.9841472
var.test(assayA3, assayB3)
> var.test(assayA3, assayB3)
F test to compare two variances
data: assayA3 and assayB3
F = 3.4223, num df = 19, denom df = 19, p-value = 0.01016
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
1.354598 8.646337
sample estimates:
ratio of variances
3.422325
This time we detect a significant difference in the variance of the two assays, with p=0.01. We have
successfully reduced the variance of the assay, and have demonstrated the reduction is statistically
significant. Reducing the variance of the assay should help make all our future experiments more
powerful, because we have reduced the unexplained variance.
Population variance versus sample variance in small samples
The previous example showed that we may have very low power to detect a reduction of variance when
we use a small sample size. Sample variance estimated using small sample sizes may also give us quite
misleading estimates of the population variance. Here's an example.
We'll generate samples from a normal distribution that has mean=10, sd=2, using the R function
rnorm(n,mean=10,sd=2). Notice that sd=2, so variance=sd2 = 22 = 4.
The samples we generate will have sample sizes ranging from 3 to 29. We'll generate 100 samples of size
n=3. We'll calculate and plot the variances of each of those 100 samples. Then we'll repeat the
operations for samples of size n from 3 to 29 in steps of 2.
plot(c(0,30),c(0,20),type="n",xlab="Sample size", ylab="Variance")
abline(h=4)
for(n in seq(3,29,2))
{for(I in 1:100)
{x=rnorm(n,mean=10,sd=2)
points(n,var(x))
}}
The plot shows sample size versus variance for the 100 samples at each sample size. The horizontal line
is the true population variance = 4. Notice that the sample variances can be quite different from the true
population variance. In particular, notice what happens when the sample size is less than 10. It is easy to
be misled about variance when we use small sample sizes. For sample sizes of 20 and above, we don't
see such extreme sample variances.
F statistic = the ratio of two variances
In a previous example, we used var.test() to test if the variances of two samples were significantly
different. The var.test() function is based on the F statistic, which is the ratio of the two variances.
Several statistical tests are based on the F statistic. For an analysis of variance or regression analysis you
may have seen the F statistic reported for the overall (omnibus) null hypothesis that none of the
independent variables were related to the dependent variable. For the special case of a t-test, the
squared T statistic equals the F statistic, that is T2 = F. Because it is widely used, you may find it useful to
understand the F statistic a little better, so we'll examine it here.
We'll start with our two assays, assayA and assayB. Suppose that, for this example, the null hypothesis is
true. That is, the two assays have the same population mean=20, and the same population standard
deviation=2, giving the same population variance=4. Suppose we generate a sample of size n=20 for
assayA and a sample of size n=20 for assayB, and then calculate the ratio of their sample variances, that
is we calculate F = var(assayA)/var(assayB). What would the distribution of the F statistic look like, when
the null hypothesis is true?
Here is R code to generate the distribution of the F statistic when the null hypothesis is true, with n=20
observations per group.
n = 20
var.ratio.list = c()
for (i in 1:10000)
{
assayA.sample = rnorm(n, mean=20, sd=2)
assayB.sample = rnorm(n, mean=20, sd=2)
var.ratio.list[i] = var(assayA.sample)/var(assayB.sample)
var.ratio.list
}
# Plot the var.ratio.list
hist(var.ratio.list, xlim= c(0, ceiling(max(var.ratio.list))), breaks=seq(0,20,.1))
In these 10,000 samples, when the null hypothesis is true, with n=20 observations per group, what are
the smallest and largest F values we see?
min(var.ratio.list)
[1] 0.1622086
max(var.ratio.list)
[1] 7.851428
The R function summary() can give us more information about the distribution of F values.
summary(var.ratio.list)
Min. 1st Qu.
0.1622 0.7404
Median
1.0130
Mean 3rd Qu.
1.1290 1.3780
Max.
7.8510
In the statistics literature, the values produced by the summary() function are called quantiles of the
distribution.
The R function quantile() can give us more details.
To get the minimum value in var.ratio.list, we use this command.
quantile(var.ratio.list,probs=0.0)
0%
0.1622086
The "0%" in the output tells us that 0% of the values are less than 0.1622086. It is the minimum value.
To get the maximum value in var.ratio.list, we use this command.
quantile(var.ratio.list,probs=1.0)
100%
7.851428
The "100%" in the output tells us that 0% of the values are less than 7.851428. It is the minimum value.
What would this command give us?
quantile(var.ratio.list,probs=0.5)
50%
1.013014
The "50%" in the output tells us that 50% of the values are less than 1.01304. So 1.01304 is the median
value.
Let's collect up several quantiles of interest.
mylist=c(quantile(var.ratio.list,probs=0.0),
quantile(var.ratio.list,probs=0.25),
quantile(var.ratio.list,probs=0.5),
quantile(var.ratio.list,probs=0.75),
quantile(var.ratio.list,probs=1))
mylist
> mylist
0%
25%
50%
75% 100%
0.1622086 0.7404088 1.0130136 1.3778022 7.8514283
Using quantiles() gives us another (more flexible) way to can get the same information as we get from
summary()
summary(var.ratio.list)
Min. 1st Qu.
0.1622 0.7404
Median
1.0130
Mean 3rd Qu.
1.1290 1.3780
Max.
7.8510
So why would we use quantiles? Because we are interested in the quantile corresponding to the 95th
percentile.
quantile(var.ratio.list,probs=0.95)
95%
2.175191
The output tells us that, when the null hypothesis is true (the two groups have the same variance), for
n=20 per group, 95% of all the F-statistics are less than 2.175. Let's think back to the example where we
wanted to test if assayB had lower variance than assayA. We take a sample for assayA, a sample for
assayB, and we calculate the F statistic for the ratio of their variances, F = var(assayA)/var(assayB). Then
we compare the F statistic for the actual assayA and assayB to the F distribution that we just generated.
> assayA3=rnorm(n=20, mean=20, sd=2)
> assayA3
[1] 20.23055 22.28764 20.47531 22.38541 21.89745 22.69879 20.26005 18.21365 16.79730 18.70380
[11] 16.67199 20.02466 22.60145 18.22082 20.03363 19.28665 20.80830 19.43308 21.02003 21.69179
> assayB3=rnorm(n=20, mean=20, sd=1)
> assayB3
[1] 18.44107 20.54730 21.32686 19.32063 19.38746 19.99892 20.85614 20.29317 21.89105 19.31109
[11] 19.98739 20.12810 21.60681 21.80682 19.12465 20.79461 18.71171 20.15293 20.27934 20.09736
>
> sd(assayA3)
[1] 1.820626
> sd(assayB3)
[1] 0.9841472
var(assayA3)
var(assayB3)
F.forA3B3 = var(assayA3)/var(assayB3)
F.forA3B3
> var(assayA3)
[1] 3.314679
> var(assayB3)
[1] 0.9685457
> F.forA3B3 = var(assayA3)/var(assayB3)
> F.forA3B3
[1] 3.422325
So for this example, the actual F for the ratio of their variances, F = var(assayA)/var(assayB), is F =
3.422325.
The actual F =3.422325 is greater than 2.175 (the 95th percentile of the F distribution under the null
hypothesis), so we can make the following statement.
If the null hypothesis is true, the probability that we would see as extreme a ratio of the two group
variances is less than 5%. That is, p < 0.05.
Our observed F-statistic of 3.422 is actually greater than 99% of the F values in our simulation under the
null distribution. So we could say we have p < 0.01.
quantile(var.ratio.list,probs=0.99)
99%
3.086891
Here is the result we got when we used the R function var.test().
var.test(assayA3, assayB3)
> var.test(assayA3, assayB3)
F test to compare two variances
data: assayA3 and assayB3
F = 3.4223, num df = 19, denom df = 19, p-value = 0.01016
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
1.354598 8.646337
sample estimates:
ratio of variances
3.422325
Notice that the value of F=3.4223 is the same as the ratio of the variances, which we also calculated. The
p-value is p=0.01016, which is slightly different from what we got with our simulation. If we used a
larger number of replicates in our simulation (more than 10,000), our p-value would converge to
p=0.01016, which is derived from the theoretical F distribution. The figure below shows the histogram of
values of the F statistic from 1000 simulations and from the theoretical F distribution.
hist(var.ratio.list, xlim= c(0, ceiling(max(var.ratio.list))), breaks=seq(0,20,.1), xlab="F statistic =
var(assayA)/var(assayB)", main="Histogram of F statistics from simulation using null hypothesis \n and
theoretical F distribution")
xvalues=seq(0,8,0.01)
yvalues=df(xvalues,19,19)*1000
yvalues
lines(xvalues, yvalues, col=2)
qf(0.95,19,19)
[1] 2.168252
pf(3.4223,19,19)
[1] 0.9949207
2*(1- pf(3.4223,19,19))
[1] 0.01015858