tpj12627-sup-0012-Legends
advertisement

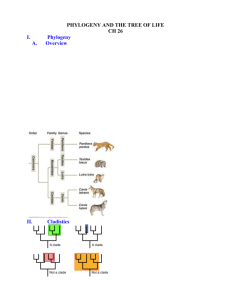
SUPPORTING INFORMATION Additional Supporting Information may be found in the online version of this article. Figure S1. Composition of the 432 candidate gap-filling sets of 44 reactions restoring the producibility of 50 of the 51 target metabolites. Each set can be decomposed into 35 ubiquitous reactions, plus one reaction in each clique, together with two reactions from the last group (top left). Figure S2. Phylogenetic analysis of the E. siliculosus chorismate mutase. For each sequence, the first part of the label corresponds to the NCBI accession number and the second part gives the species in which this sequence is retrieved. The color indicates the phylogenetic origin of the species considered. Figure S3. Phylogenetic analysis of the E. siliculosus prephenate aminotransferase (PAT). For each sequence, the first part of the label corresponds to the NCBI accession number and the second part gives the species in which this sequence is retrieved. The color indicates the phylogenetic origin of the species considered. Figure S4. Phylogenetic analysis of the E. siliculosus arogenate/prephenate dehydratase (A/P-DT). For each sequence, the first part of the label corresponds to the NCBI accession number and the second part gives the species in which this sequence is retrieved. The color indicates the phylogenetic origin of the species considered. Figure S5. Phylogenetic analysis of the E. siliculosus arogenate/prephenate dehydrogenase (A/P-DH). For each sequence, the first part of the label corresponds to the NCBI accession number and the second part gives the species in which this sequence is retrieved. The color indicates the phylogenetic origin of the species considered. Figure S6. Phylogenetic analysis of the E. siliculosus aromatic amino acid aminotransferase (AA-AT). For each sequence, the first part of the label corresponds to the NCBI accession number and the second part gives the species in which this sequence is retrieved. The color indicates the phylogenetic origin of the species considered. Figure S7. Phylogenetic analysis of Phe hydroxylases (PAHs). The text line corresponds either to the phylum, to the main eukaryotic/prokaryotic group or to the species name. When the branch represents several species and was collapsed, the number of species is mentioned into brackets. Only bootstrap values >60 are indicated. Table S1. List of compounds known to be produced by E. siliculosus under normal growth conditions and considered as targets for testing the metabolic capacity of EctoGEM. Values corresponding to the targets used to build the biomass function are provided in µmol/g DW. Table S2. List of the 35 reactions from MetaCyc necessary to restore the producibility of target metabolites. Table S3. List of pathways that have been manually validated within EctoGEM. Table S4. List of E. siliculosus genes reannotated in the Orcae database based on information gained through EctoGEM.