AWC_Logan_PhyloNetworks
advertisement
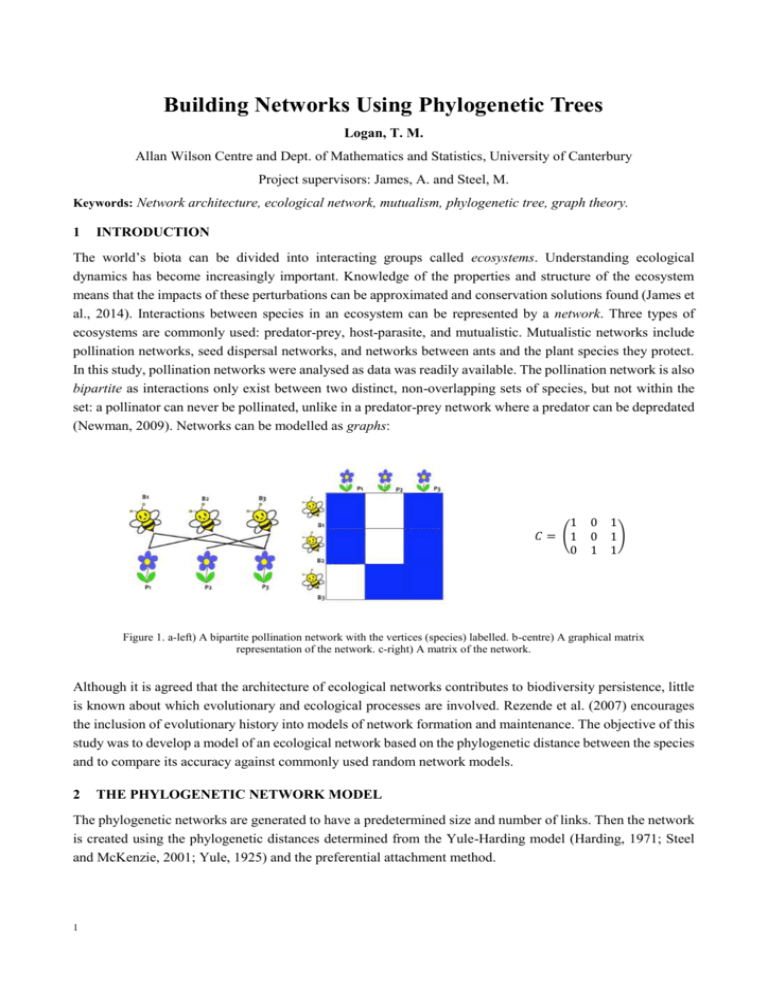
Building Networks Using Phylogenetic Trees Logan, T. M. Allan Wilson Centre and Dept. of Mathematics and Statistics, University of Canterbury Project supervisors: James, A. and Steel, M. Keywords: Network architecture, ecological network, mutualism, phylogenetic tree, graph theory. 1 INTRODUCTION The world’s biota can be divided into interacting groups called ecosystems. Understanding ecological dynamics has become increasingly important. Knowledge of the properties and structure of the ecosystem means that the impacts of these perturbations can be approximated and conservation solutions found (James et al., 2014). Interactions between species in an ecosystem can be represented by a network. Three types of ecosystems are commonly used: predator-prey, host-parasite, and mutualistic. Mutualistic networks include pollination networks, seed dispersal networks, and networks between ants and the plant species they protect. In this study, pollination networks were analysed as data was readily available. The pollination network is also bipartite as interactions only exist between two distinct, non-overlapping sets of species, but not within the set: a pollinator can never be pollinated, unlike in a predator-prey network where a predator can be depredated (Newman, 2009). Networks can be modelled as graphs: 1 𝐶 = (1 0 0 0 1 1 1) 1 Figure 1. a-left) A bipartite pollination network with the vertices (species) labelled. b-centre) A graphical matrix representation of the network. c-right) A matrix of the network. Although it is agreed that the architecture of ecological networks contributes to biodiversity persistence, little is known about which evolutionary and ecological processes are involved. Rezende et al. (2007) encourages the inclusion of evolutionary history into models of network formation and maintenance. The objective of this study was to develop a model of an ecological network based on the phylogenetic distance between the species and to compare its accuracy against commonly used random network models. 2 THE PHYLOGENETIC NETWORK MODEL The phylogenetic networks are generated to have a predetermined size and number of links. Then the network is created using the phylogenetic distances determined from the Yule-Harding model (Harding, 1971; Steel and McKenzie, 2001; Yule, 1925) and the preferential attachment method. 1 Figure 2. The mutualistic pollination network. The red dashed line is the seed link between the plants and pollinators. The other links are weighted (width) to show the likelihood of a connection being formed between them. 3 DISCUSSION The phylogenetic network model was refined by varying the input parameters. Then, for each real set of network data and the corresponding generated model, the metrics were compared. The phylogenetic network performed better than the random network in all metrics, except of modularity. This is likely due to the phylogenetic networks having an accurate nestedness, and the data collection method (of the networks which the models were evaluated on) favoured modular network by excluding outlying species. One of the network’s compiling author: “… in this initial survey we limit ourselves to relative importance of major vector types…” (Arroyo et al., 1982) (p.84) A further application for this work is to investigate the implications of a phylogenetic model on the study of community evolution. It may be possible to determine genetic similarity based on species interactions, or determine whether a currently unknown species should have existed by examining gaps in network interactions. Further work is to determine the suitability of the phylogenetic model for networks other than binary, bipartite, pollination networks. Ultimately, the model may provide further insight into ecosystem responses to perturbations and contribute to the conservation of ecological networks. 4 ACKNOWLEDGEMENTS Thank you to the Allan Wilson Centre for providing funding for this project. 5 REFERENCES Arroyo, M. T. K., Primack, R., and Armesto, J. (1982). "Community studies in pollination ecology in the high temperate Andes of central Chile. I. Pollination mechanisms and altitudinal variation." American journal of botany, 82-97. Harding, E. (1971). "The probabilities of rooted tree-shapes generated by random bifurcation." Advances in Applied Probability, 44-77. James, A., McLeod, J., and Pitchford, J. (2014). "Simple Cycles Underpin Ecological Complexity." Unpublished manuscript. Newman, M. (2009). Networks: an introduction, Oxford University Press. Rezende, E. L., Lavabre, J. E., Guimarães, P. R., Jordano, P., and Bascompte, J. (2007). "Non-random coextinctions in phylogenetically structured mutualistic networks." Nature, 448(7156), 925-928. Steel, M., and McKenzie, A. (2001). "Properties of phylogenetic trees generated by Yule-type speciation models." Mathematical biosciences, 170(1), 91-112. Yule, G. U. (1925). "A mathematical theory of evolution, based on the conclusions of Dr. JC Willis, FRS." Philosophical Transactions of the Royal Society of London. Series B, Containing Papers of a Biological Character, 213, 2187. T. Logan