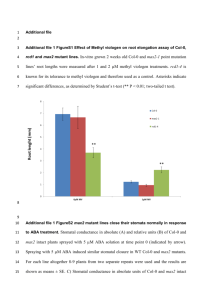
tpj13118-sup-0005-Legend
advertisement

Supporting Information Legends Figure S1. Confirmation of dog1-1 microarray data by RT-qPCR. The identified expression differences between NILDOG1 and dog1-1 were further investigated by RT-qPCR in four genotypes, including a second dog1 allele (dog1-3, which is a SALK_000867 T-DNA insertion mutant) and its corresponding wild type Columbia (Col-0). We confirmed the expression of several genes that were down regulated (EARLY METHIONINE-LABELLED (EM) 1, EM6, ABI5 and ABRE BINDING FACTOR (ABF) 4), not changed (G-BOX BINDING FACTOR (GBF) 4) as well as several genes that were found to be enhanced (ABI4, EXPANSIN A2, and ENDO-ß-MANNANASE (MAN) 7) in the dog1-1. The relative expression in dog1-1 (compared with NILDOG1, set to 1) of the genes in the microarray (top row), by RT-qPCR in dog1-1 (compared with NILDOG1, set to 1) and RT-qPCR in dog1-3 (compared with Col-0, set to 1). The green color represents reduced expression compared with WT and red an enhanced expression in dog1-1. The asterisks indicate that the expression in dog1 is significantly different from WT (Student’s t test, p<0.05). nd = not detected. Figure S2. Seed sugar content of the dog1-1 abi3-1 double mutant mimics that of the severe abi3-5 mutant. The graphs show the content of (a) sucrose, (b) glucose and fructose and (c) maltose, xylose, myo-inositol, galactinol, raffinose, stachyose and trehalose in six different genotypes. The green seeded double mutant dog1-1 abi3-1 and the green seeded abi3-5 mutant are characterized by increased amounts of sucrose, glucose, fructose, maltose, xylose, myo-inositol, while the compounds of the RFO pathway are strongly decreased. The values are means ± SEM of three biological replicates. Figure S3. Proteome analysis of the dog1 and abi3 mutants. (a) The list shows the 12 protein that differentially accumulated in dog1-1 (fold change >2, p<0.05). (b) Fold change difference of five seed storage proteins as compared with Ler-0. (c) Fold change difference of 7 OLEOSIN proteins as compared to Ler-0. Figure S4. Overlap between up-regulated gene sets in abi5-7 and dog1-1 seeds. (a) Venn-diagram showing the overlap of genes up-regulated in both dog1-1 and abi5-7 mutant seeds. (b) As a control we also investigated the overlap between up-regulated set in dog1-1 with the down-regulated set in abi5-7. Figure S5. Microarray quality and reproducibility. (a,b) The data from the 9 arrays used showed after hybridization similar patterns of intensity. Slide hybridization patterns were inspected manually as well without detecting physical or hybridization artifacts (data not shown). The RNAs used as templates for probe synthesis were shown to be intact based on Bio-Analyzer 2001 analysis. (c) In agreement with this were the hybridization patterns of control genes on the slides showing a near-identical pattern of hybridization over the mRNA lengths, which indicate similar RNA and probe integrity. (d,e) The uniformity of normalized unscaled SE (NUSE) and relative log expression (RLE) indicate high quality and uniformity of the hybridization data, with one slide showing a small deviation (Brettschneider et al., 2008). (f,g) Raw intensity data were subjected to RMA normalization (Irizarry et al., 2003), which kept the uniformity of general levels between the different slides. (h) Between replicate reproducibility of the experiment was high, exemplified by the high correlation between the data of two representative biological replicates. Table S1. List of genes related to ABA signalling that are down-regulated in dog1-1 mutant seeds. Table S2. Late embryogenesis abundant and heat shock protein genes that are down-regulated in dog1-1. Table S3. Overview of 13 genes whose expression level was tested by RT-qPCR in abf4, abi5, dog1, abi5 dog1, abi5 abf4 and abi5 dog1 abf4 mutants. Table S4. Primers sets used for genotyping ABF4 and DOG1 T-DNA insertion mutants on the mRNA level. Table S5. Primer sets used for RT-qPCR. Methods S1. Detailed description of the metabolite analysis by GC-TOF-MS and protein quantification and identification. Data S1. Sets of differentially expressed genes in dog1-1 mutant seeds.