Electronic Supplementary Material
advertisement

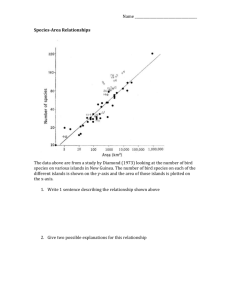
Electronic Supplementary Material Historical species losses in bumblebee evolution Fabien L. Condamine and Heather M. Hines Contents Text S1 Extended Methods. Table S1 Time-dependent diversification analyses. Figure S1 Convergence of the BAMM analysis. Figure S2 Frequency distribution of distinct macroevolutionary rate regimes estimated using BAMM. Figure S3 Credible set of configuration shifts of bumblebees inferred with BAMM. Figure S4 Estimates of speciation rates along the bumblebee phylogeny obtained from BAMM analyses. Figure S5 Estimates of extinction rates along the bumblebee phylogeny obtained from BAMM analyses. Text S1. Extended Methods Diversification analyses While time-constant models are useful, there are many reasons why diversification rates can vary over the evolutionary history of a clade, including changes in the biotic and abiotic environments, diversity-dependent effects, or the combination of both. A straightforward and widespread approach to account for time variation in diversification rates is to assume a functional dependence of speciation and extinction rates with time [1-5]. Likelihood expressions of phylogenetic branching times for such models are available, for both continuous (e.g. linear, exponential; [2,4,5]) and discrete (referred to as the ‘discrete shift’ model; [3,5]) forms of time variation. These time-dependent models allow a quantitative estimation of how diversification rates have varied through time. Discrete time-variation of diversification The TreePar package [3] was used to assess speciation and extinction rates through time. This method relaxes the assumption of constant rates by allowing rates to change at specific points in time. Such a model allows for the detection of rapid changes in speciation and extinction rates due to environmental factors like the geological changes in the region. We employed the ‘bd.shifts.optim’ function that allows for estimating discrete changes in speciation and extinction rates and mass extinction events in under-sampled phylogenies. Going backward in time, it estimates the maximum likelihood speciation and extinction rates together with the rate shift times t=(t1,t2,...,tn) in a phylogeny. At each time t, the rates are allowed to change and the species may undergo a shift in diversification. TreePar analyses were run with the following settings: start = 0, end = crown age estimated by dating analyses, grid = 0.1 Myr, four possible shift times were tested, and posdiv = FALSE to allow the diversification rate to be negative (i.e. allows for periods of declining diversity). TreePar analyses were run as follows: start=0 (present), end=Bombus crown age (34 million years, Myr, see Hines [2008]), grid=0.1 Myr (examining potential rate shifts every 0.1 Myr), with no more than six shift times. Speciation and extinction rates are inferred given the diversification rate (=speciation–extinction) and turnover (=extinction/speciation). Continuous time-variation diversification We used the Morlon et al. [4]’s approach. This method has the advantage to take into account the heterogeneity of diversification rates across the tree such that clades may have their own speciation and extinction rates (and their own diversity dynamics), and to estimate continuous variations of rates over time (whereas extinction is constant in BAMM). The Morlon et al.’s approach is suitable to potentially infer a declining diversity pattern in which extinction can exceed speciation meaning that diversification rates can be negative [4]. We designed four models to be tested: (i) BCSTDCST, speciation and extinction rates are constant; (ii) BVARDCST, speciation rate is exponentially varying and extinction rate is constant; (iii) BCSTDVAR, speciation rate is constant and extinction rate is exponentially varying; and (iv) BVARDVAR, speciation and extinction rates are exponentially varying. We also repeated these four models with a linear dependence through time. Models with exponential variation are: 𝜆(𝑡) = 𝜆0 × 𝑒 𝛼𝑡 and 𝜇(𝑡) = 𝜇0 × 𝑒 𝛽𝑡 , and models with linear variation are: 𝜆(𝑡) = 𝜆0 + 𝛼𝑡) and 𝜇(𝑡) = 𝜇0 + 𝛽𝑡), in which 𝜆0 and 𝜇0 are the speciation and extinction rates at present, and α and β are the rates of change according to time. Across-clade and time-variation diversification We used the Bayesian Analysis of Macroevolutionary Mixture (BAMM, www.bammproject.org) to estimate speciation and extinction rates through time and among/within clades [6]. BAMM is an analytical tool for studying complex evolutionary processes on phylogenetic trees, potentially shaped by a heterogeneous mixture of distinct evolutionary dynamics of speciation and extinction across clades. The method uses reversible jump MCMC (rjMCMC) to detect automatically rate shifts and sample distinct evolutionary dynamics that best explain the whole diversification dynamics of the clade. Within a given regime, evolutionary dynamics can involve time-variable diversification rates; in BAMM, the speciation rate is allowed to vary exponentially through time while extinction is maintained constant [7]. Subclades in the tree might diversify faster (or slower) than others, and BAMM allows detecting these diversification rate shifts without a priori hypotheses on how many and where these shifts might occur. BAMM provides estimates of marginal probability of speciation and extinction rates at any point in time along any branch of the tree. Marginal probabilities of the number of evolutionary regimes can also be computed, allowing comparisons of models with a given number of shifts with Bayes factors. BAMM is implemented in a C++ command line program and the BAMMtools R-package [8]. We ran BAMM analyses on the chronogram calibrated with four fossils, and reconstructed with either the Yule or the birth-death prior. We set four rjMCMC running for 507 generations and sampled every 50,000 generations. We accounted for incomplete taxon sampling using the implemented analytical correction, with a sampling fraction taking into account the missing species. We performed four independent runs (with a burn-in of 10%) using different seeds, and we used ESS to assess the convergence of the runs, considering values above 200 as indicating good convergence. The posterior distribution was used to estimate the configuration of the diversification rate shifts; alternative diversification models were compared using Bayes factors. Model selection For TreePar and the Morlon et al.’s approach, we computed the corrected Akaike information criterion (AICc) based on the log-likelihood and the number of parameters of each diversification model [9]. A difference of two in the AICc (ΔAICc) between two models suggests significant support for the model with the lowest AICc. We also checked support for the selected model against all models nested within it using the likelihood ratio test (LRT), which shows significant support at P < 0.05. The scenario supported by LRT and with the lowest AICc was considered the best. If the model with the lowest AICc was not supported by LRT, the model with less parameter was considered the best. For the BAMM analyses, we used posterior probabilities and Bayes factors to compare the fit of different evolutionary scenarios (i.e. with no shift, one shift…). We considered Bayes factors values above 5 to significantly favour one model over another [10]. References 1. Nee S, May RM, Harvey PH. 1994b The reconstructed evolutionary process. Philos. Trans. R. Soc. Lond. B Biol. Sci. 344: 305 – 311. (doi.org/10.1098/rstb.1994.0068) 2. Rabosky DL, Lovette IJ. 2008 Explosive evolutionary radiations: decreasing speciation or increasing extinction through time? Evolution 62: 1866–1875. (doi.org/10.1111/j.1558-5646.2008.00409.x) 3. Stadler T. 2011 Mammalian phylogeny reveals recent diversification rate shifts. Proc. Natl. Acad. Sci. USA 108, 6187–6192. (doi.org/10.1073/pnas.1016876108) 4. Morlon H, Parsons TL, Plotkin J. 2011 Reconciling molecular phylogenies with the fossil record. Proc. Natl Acad. Sci. USA 108, 16327–16332. (doi.org/10.1073/pnas.1102543108) 5. Hallinan N. 2012 The generalized time variable reconstructed birth–death process. J. Theor. Biol. 300: 265–276. (doi.org/10.1016/j.jtbi.2012.01.041) 6. Rabosky DL, Santini F, Eastman JM, Smith SA, Sidlauskas B, Chang J, Alfaro ME. 2013 Rates of speciation and morphological evolution are correlated across the largest vertebrate radiation. Nature Commun. 4, 1958. (doi.org/10.1038/ncomms2958) 7. Rabosky DL, Donnellan SC, Grundler M, Lovette IJ. 2014 Analysis and visualization of complex macroevolutionary dynamics: An example from Australian scincid lizards. Syst. Biol. 63: 610-627. (doi.org/10.1093/sysbio/syu025) 8. Rabosky DL, Grundler M, Anderson C, Shi JJ, Brown JW, Huang H, Larson JG. 2014b BAMMtools: an R package for the analysis of evolutionary dynamics on phylogenetic trees. Methods Ecol. Evol. 5: 701–707. (doi.org/10.1111/2041-210X.12199) 9. Cavanaugh JE. 1997 Unifying the derivations of the Akaike and corrected Akaike information criteria. Stat. Prob. Lett. 31: 201–208. (doi.org/10.1016/S01677152(96)00128-9) 10. Kass RE, Raftery AE. 1995 Bayes factors. J. Am. Stat. Assoc. 90: 773-795. (doi.org/10.1080/01621459.1995.10476572) Table S1. Time-dependent diversification analyses. The TreePar model with four shifts, and the model with speciation and extinction varying linearly though time (BTimeVarDTimeVar_LIN) are supported. (a) TreePar models, in which ‘r1’ denotes the diversification rate and ‘τ1’ is the turnover, both inferred between Present and the shift time 1 (‘st1’). (b) Models of Morlon et al., in which λ0 and μ0 are the speciation and extinction rates at present, with α and β the rates of their respective variation through time. (a) Models Constant birth-death BD with 1 shift BD with 2 shifts BD with 3 shifts BD with 4 shifts N logL -460.44 ±2.366 -449.46 5 ±2.478 -445.57 8 ±2.500 -438.55 11 ±2.506 -434.44 14 ±2.501 2 AICc 924.87 ±4.733 908.92 ±4.956 907.15 ±5.000 899.10 ±5.012 896.88 ±5.002 ΔAICc AIC weight P (LRT) 27.99 0 - 12.03 0.0013 <0.0001 10.26 0.00 0.051 2.214 0.17 0.001 0 0.515 0.042 ΔAICc AIC weight P (LRT) r1 τ1 st1 r2 τ2 st2 r3 τ3 st3 r4 τ4 st4 r5 τ5 0.175 ±0.001 0.032 ±0.003 -0.217 ±0.004 -1.538 ±0.006 -2.036 ±0.005 0.000 ±0.008 0.409 ±0.016 5.017 ±0.030 28.719 ±0.054 28.330 ±0.054 - - - - - - - - - - - - 0.8 ±0.039 0.8 ±0.046 0.8 ±0.044 0.8 ±0.040 0.155 ±0.001 0.100 ±0.020 0.561 ±0.032 0.679 ±0.037 - - - - - - - - - - - - - λ0 α μ0 β - - - - 0 ±0.0051 - - - 0 ±0 - (b) Models N Yule model 1 Constant birth-death 2 BTimeVar_EXPO 2 BTimeVarDCST_EXPO 3 BCSTDTimeVar_EXPO 3 BTimeVarDTimeVar_EXPO 4 BTimeVar_LIN 2 BTimeVarDCST_LIN 3 BCSTDTimeVar_LIN 3 BTimeVarDTimeVar_LIN 4 logL AICc -610.86 ±2.512 -610.86 ±2.631 -610.63 ±2.708 -610.63 ±2.708 -610.86 ±2.706 -610.63 ±2.731 -610.58 ±2.679 -610.58 ±2.695 -609.97 ±2.702 -600.79 ±2.633 1223.741 ±5.025 1225.778 ±5.261 1225.316 ±5.415 1227.373 ±5.415 1227.835 ±5.413 1229.448 ±5.463 1225.213 ±5.357 1227.269 ±5.391 1226.048 ±5.405 1209.778 ±5.267 0.1742 ±0.0025 0.1742 16.000 0.0003 0.99 ±0.0051 0.1823 15.538 0.0004 0.497 ±0.0041 0.1822 17.595 0.0001 0.794 ±0.0041 0.1743 18.057 0.0001 0.99 ±0.0044 0.1821 19.670 0.00005 0.927 ±0.0041 0.1837 15.435 0.0004 0.453 ±0.0032 0.1838 17.491 0.0002 0.754 ±0.0040 0.1833 16.270 0.0003 0.409 ±0.0041 0.0272 0 0.997 <0.0001 ±0.0056 13.963 0.0009 - -0.0077 ±0.0011 -0.0077 ±0.0011 0 ±0.0029 -0.0076 0 ±0.0016 ±0.0005 -0.0016 ±0.0002 -0.0016 0 ±0.0002 ±0.0010 0.0079 ±0.0018 0.1353 0.0814 ±0.0029 ±0.0021 - 0.0187 ±0.0104 0.0256 ±0.0088 0.0040 ±0.0005 -0.1291 ±0.0040 0.345 ±0.010 0.664 5.6 ±0.094 ±0.761 0.000 3.3 ±0.114 ±0.535 0.000 3.3 ±0.121 ±0.486 0.102 ±0.011 -0.188 ±0.027 -0.237 ±0.148 0.826 ±0.036 1.746 5.6 ±0.040 ±0.748 1.731 5.6 ±0.042 ±0.589 0.106 ±0.038 -0.002 ±0.031 0.892 ±0.034 1.002 14.6 0.050 0.991 ±0.115 ±0.697 ±0.153 ±0.030 Figure S1. Convergence of the BAMM analysis with the chronogram. (a) The stationary of the MCMC before applying a burn-in. (b) The posterior distribution of number of shifts estimated before applying a burn-in. (c) The stationary of the MCMC after removing the burn-in phase. (d) The posterior distribution of number of shifts estimated after removing the burn-in phase. Figure S2. Frequency distribution of distinct macroevolutionary rate regimes estimated using BAMM. (a) Prior distribution of the number of distinct processes. (b) Posterior distribution of the number of distinct processes (including the root process). A one-process model outperforms a two-process model. Figure S3. Credible set of configuration shifts of bumblebees inferred with BAMM. For each shift configuration, the locations of rate shifts are shown as red (rate increases) and blue (rate decreases) circles, with circle size proportional to the marginal probability of the shift. Text labels (e.g. f =0.86) denote the posterior probability of each shift configuration. Figure S4. Estimates of speciation rates along the bumblebee phylogeny obtained from BAMM analyses. Colours at each point in time along branches denote instantaneous rates of speciation inferred as the mean scenario, with colours indicating mean rates across all the shift configurations sampled in the Bayesian posterior. Figure S5. Estimates of extinction rates along the bumblebee phylogeny obtained from BAMM analyses. Colours at each point in time along branches denote instantaneous rates of extinction inferred as the mean scenario, with colours indicating mean rates across all the shift configurations sampled in the Bayesian posterior.