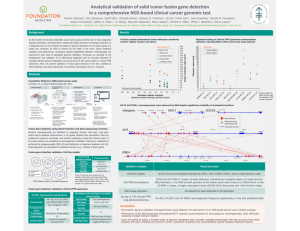
Detailed Assay Design and Validation Protocol Assay Designer 4.0
advertisement

Detailed Assay Design and Validation Protocol Assay Designer 4.0 software (Sequenom, San Diego, CA) was used to design the PCR and extension primers at the fusion points and the exon junctions based on fusion gene cDNA sequence for 15 presently identified fusion genes involving ROS1 and RET (Figure 1). Plasmids comprising synthetic fusion genes — exact cDNA surrogates for the endogenously expressed ROS1 and RET fusion genes — were used to validate the specificity and sensitivity of each assay as follows. The plasmid copy number input (~3000 plasmids/well) was optimized and PCR amplifications were run using Master Cycler Pro 384 (Eppendorf, Hauppauge, NY) in 384well plates (Thermo Scientific, Rockford, IL). The following optimum conditions were arrived at during the validation studies: 95 °C (2 minutes) followed by 45 cycles of 95 °C (30 seconds), 56 °C (30 seconds), 72 °C (60 seconds) and a final extension at 72 °C (5 minutes). Reactions per well contained 0.01 pg/well of plasmid DNA, 100nM PCR primers, 2.0 mM MgCl2, 500µM dNTPs and 1U of FS Taq polymerase (PCR enzyme kit, Sequenom) in a total 4 µL volume. To remove all excess dNTPs, the PCR products were treated with shrimp alkaline phosphatase (SAP), which dephosphorylates the unincorporated dNTPs. This step was done by adding 0.3µL SAP (1.7U/µL), 0.17µL SAP buffer and 1.53µL autoclaved deionized water to the resultant PCR with the following thermocycler parameters: 37 °C for 40 minutes and 85 °C for 5 minutes. Samples were kept at 4 °C until further use. Prior to setting up the extension PCR step, concentration adjustments were made to the extension primers by dividing them into a low mass group and a high mass groups. The initial concentrations of the low and high-mass groups were calculated to get 7 µM and 14 µM respectively. The extension reaction mixture per well contained extension buffer 0.2 µL, termination mix 0.2 µL, primer mix (low-mass = 0.625 µM and high mass = 1.25 µM) in 0.804 µL, thermosequenase extension enzyme 0.041 µL (1.35 units / reaction) and 0.755 µL sterile water. The extension reaction conditions were as follows: 94 °C (30 seconds) followed by 40 cycles of 94 °C (5 seconds), 5 cycles of 52 °C (5 seconds), 80 °C (5 seconds) and a final extension step of 72 °C (3 minutes). The reaction products were conditioned with a cationic exchange resin (Clean resin) to remove excess salts (Na+, K+) to minimize background noise.15-20 nano liters of the analyte (based on the volume check) were spotted onto the MassARRAY spectroCHIP(r) II with a RS1000 Nanodispenser and the chip was loaded into the Mass spectrometer (Sequenom). Data Analysis MassARRAY Typer v 4.0.53 software (Sequenom, San Diego, CA) was used to assemble data from mass spectrometry analysis. The accurate detection of a fusion gene (or lack thereof) in our assay considers a number of observations. The presence of expected peaks from both directions (forward and reverse) at a fusion point confirms that the fusion is present. If there is no peak from assaying the fusion junction from both directions and the presence of mass spectrometry peaks corresponding to upstream and downstream wild-type gene exon junctions are observed, the absence of the fusion is confirmed. A false negative result would be indicated if wild-type exon junctions and fusion junction peaks are not detected. A potential false positive result is indicated if the fusion gene junction is only detected by one of the two targeted fusion gene extension primers.