C483 Discussion Week of January 14 “The Crystal Structures of
advertisement

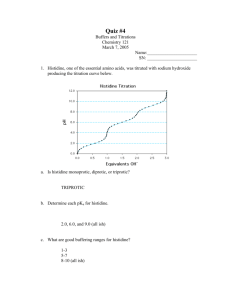
C483 Discussion Week of January 14 “The Crystal Structures of SARS Virus Main Protease and Its Complex with an Inhibitor” http://www.pnas.org/content/100/23/13190 Names: 1. _____________________________________ 2. ____________________________________ 3. _____________________________________ 4. ____________________________________ Questions: Part 1: Enzyme activity Assay. The investigators needed a way to test how active Main Protease was at different pHs. They used a substrate, which is the reactant in the enzyme catalyzed reaction. After the reaction ran for a certain time, they used analytical methods to determine how much product had been formed relative to how much of the substrate was unreacted. 1. (2pts) Main Protease is specific for hydrolyzing proteins and peptides on the carboxy end of Gln. The following substrate was used to test activity: Thr-Ser-Ala-Val-Leu-Gln-Ser-Gly-Phe-Arg Draw the chemical structure of the substrate, with correct stereochemistry and predominant ionization, at pH 8. Then draw the structures of the hydrolysis products at pH 8. Do the ionization states of any of these structures differ at pH 6? 2. (2pts) The rate of reaction was determined by HPLC for these enzyme assays, and a spectrum like Fig 3B from the paper was obtained. A. Why are there three peaks on the graph? What structures correspond to P1, P2, and S? B. From the graph, estimate the % completion of the reaction that was analyzed. Explain how you did this. C. If the same reaction were run again, but was allowed to run for a longer time, the HPLC would change. Based on Figure 3B, draw the HPLC profile you would expect to see for this longer reaction. Part 2: Structural studies. Crystallography is a technique by which we can obtain a 3-D picture of the structure of the protein, including how it is folded up on itself, which amino acid residues interact with each other, and which interact with substrates. In this paper, the protein structure was obtained for four conditions: at pH 6, pH 7.6, pH 8.0, and pH6.0 with inhibitor. At pH 7.6 and pH 8.0, the structure was that of the active enzyme (called Protomer A). At pH 6, the protein was a mixture of active Protomer A, and inactive Protomer B. Their structures are different, and their amino acids interact differently. Therefore, when the paper talks about “His-A163” it means that this is a histidine residue in the 163 position, which is in the active structure. “His-B163” is the corresponding histidine residue in the inactive structure—the same residue, but it interacts differently with other residues in this inactive state. 3. (1pt) Which residue is protonated at pH 6 which is not protonated at pH 8? Does this protonation lead to activation or inactivation of the enzyme? What amino acid interaction is broken when protonation occurs? (1 pt) Draw a schematic of “normal” protein interactions (Protomer A) that includes His-163, His-172, Glu-166, and Phe-140. Indicate which amino acids interaction, and the type of interaction. (2pt) Now draw a schematic of “abnormal” protein interactions (Protomer B) that includes His-163, His172, Glu-166, and Phe-140. Indicate which amino acids interaction, the type of interaction, and how this is different than “normal” interation. 4. (2pt) Refer to Figure 3A in the paper, which shows enzyme activity as a function of pH. At what pH is the enzyme fully active? At what pH is the enzyme only 10% active? Assume that most of the activity change with pH is due to protonation/deprotonation of His-163. With this model, what is the approximate pKa of His-163? Explain how you came to this conclusion. Sometimes the pKa values of sidechains of amino acids in proteins are very similar to the pKa values of free amino acids; sometimes the pKa values are very different. In this case, has the environment of the protein had little or much effect on the pKa of the Histidine sidechain relative to free amino acid histidine?