mec13038-sup-0009-AppendixS1
advertisement

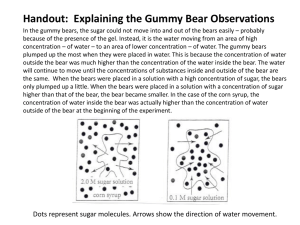
Appendix 1 Exclusion of PB7 and LS Fig. S1 describes the results of D-statistic tests (Green et al. 2010; Durand et al. 2011) for brown bear admixture into pairs of individual polar bears. Two outliers are observed: LS, which has the most positive distribution of D-statistics (indicating that the LS polar bear contains less admixture from brown bears than any other polar bear), and PB7, the only polar bear for which the distribution of D-statistics does not include zero (suggesting that PB7 may have bona fide admixture from brown bears). We suspect that both of these results are artefacts resulting from errors in the sequencing libraries rather than admixture with brown bears. As reported previously (Cahill et al. 2013), the LS polar bear data contain DNA damaged sites. This ancient DNA associated error biases the D-statistic away from zero. The LS data were generated from a 40-year-old bone that is currently stored at the National Museum of National History in Washington, DC (Cahill et al. 2013). These data show nearly twice the number of C to T and G to A transitions when compared to the polar bear reference genome than the other polar bears. This is most likely due to miscoding lesions resulting from cytosine deamination to uracil, which is the most common form of ancient DNA damage (Hofreiter 2001). Although damage does not affect the D-statistic when the damaged individual is in the position of the potential introgressor, it will bias the D-statistic when the damaged sample is included in either the P1 or P2 positions (potential recipient of introgression). In this case, the effect is a false positive match between the damaged individual and the outgroup, and consequent 1 identification of any undamaged comparison individual as admixed, as seen for LS in Fig. S1. D-statistic results show that the PB7 polar bear has a stronger signal for brown bear ancestry than any other polar bear (mean D= -0.033). However, D-statistic tests involving PB7 and the Kenai brown bear (Ken) as potential introgressor are particularly extreme, with an average D of -0.066 for D(PB7, other polar bear, Ken, black bear), compared to -0.028 for D(PB7, other polar bear, other brown bear, black bear). Given that the PB7 and Ken data were generated as part of the same project (Miller et al. 2012), we explored the possibility that contamination between these data may exist. We mapped the published genomic data from PB7 and Ken to a reference polar bear mitochondrial genome (Delisle & Strobeck 2002) bwa (Li & Durbin 2010) and samtools (Li et al. 2009) under the same parameters as the analyses described in the main text (Methods). We then identified all positions in the mitochondrial genome where the consensus mitochondrial genome sequences assembled for PB7 and Ken differed from each other. At each of these positions, we then calculated the number of times the consensus allele for Ken was observed in the PB7 data set, and vice versa. For most sites, we observed zero reads in the Ken data set that matched the consensus assembly for PB7 (Fig. S2A). However, in the PB7 data set, 0.5-1.0% of PB7 reads match the Ken allele (Fig. S2A). To determine whether this result could be due to differences in sequencing depth, we down sampled the Ken and PB7 data sets to an equal number of reads, and counted the number of sites in the down sampled data sets in which the consensus allele for the opposite species was observed (Fig. S2B). We observed that as the number of reads sampled increased so did the number of Ken alleles observed in the PB7 data set. 2 These results are most simply explained by small amounts of contamination of the PB7 data set with data from Ken. We therefore chose to exclude PB7 from further analysis. Y-chromosome Analysis Recent studies have shown that the Y-chromosome haplotypes found in polar bears and form reciprocally monophyletic clades (Bidon et al. 2014). These findings include the ABC islands brown bears which at the Y-chromosome fall within the brown bear clade (Bidon et al. 2014). We assessed the same ~390 KB Y-chromosome scaffold of the polar bear reference genome (Li et al. 2011) as Bidon et al (scaffold 297) and calculated the sequence divergence between all male bears in our panel. Using the pseudohaploidization method used elsewhere in this study (Methods) we generated representative sequences for each individual at scaffold 297 and compared pairwise differences. Within polar pairwise differences ranged from 1-4 differences per 10,000 sites. Polar bear to Baranof island brown bear differences range from 11-12.5 differences per 10,000 sties. Polar bear to American black bear differences range from 29-31 differences per 10,000 sites and the Baranof brown bear has 29 differences to the American black bear per 10,000 sites. Our analytical power in this analysis is limited by only having a single male brown bear. However, insofar as our results are interpretable they show that the Baranof island brown bear (a male ABC islands bear) possesses a Y-chromosome haplotype that falls well outside the diversity of polar bear Y-chromosomes. Bidon et al observed brown bear to polar bear divergence to be ~35% of brown/polar bear divergence to black 3 bear (Bidon et al. 2014). Our observation of ~39% (Figure S3) is qualitatively consistent with previous results and probably indicative of minor filtering differences. 4 Supplemental Figures Figure S1 - D-statistic tests for brown bear admixture into individual polar bears The boxplot shows the autosomal D-statistic for each polar bear with all possible combinations of polar bear and brown bear introgressors. Negative values indicate that the individual listed on the x-axis has more polar bear ancestry. Polar bears from the Beaufort and Chukchi seas are depicted as red, those from Svalbard are blue, those from Hudson Bay are yellow, and the Lancaster Sound bear is green. Figure S2 – Tests for contamination of PB7 by Ken The frequency of reads in the PB7 and Ken data sets that may be derived from contamination by the other data set (A). To control for the detection of potential contaminant reads based on differences in coverage, we sampled fixed numbers of reads from each individual at variable sites. (B) The mean frequency of sites containing one or more potentially contaminant reads of 20 random draws of N reads. Error bars equal two standard deviations. Figure S3 – Y-chromosome pairwise difference The number of pairwise differences per site between male individuals in our panel of bears at a ~390KB Y-chromosome scaffold. We find that the Baranof sample (the only male brown bear in this study) falls outside the range of divergences observed between polar bears. The level of divergence is consistent with previous studies analysing the same scaffold with different individuals suggesting that brown bears and polar bears form reciprocally monophyletic clades at the Y-chromosome (Bidon et al. 2014). 5 References Bidon T, Janke A, Fain SR et al. (2014) Title: Brown and polar bear Y chromosomes reveal extensive male-biased gene flow within brother lineages. Molecular biology and evolution, msu109–. Cahill JA, Green RE, Fulton TL et al. (2013) Genomic evidence for island population conversion resolves conflicting theories of polar bear evolution. (MW Nachman, Ed,). PLoS genetics, 9, e1003345. Delisle I, Strobeck C (2002) Conserved primers for rapid sequencing of the complete mitochondrial genome from carnivores, applied to three species of bears. Molecular biology and evolution, 19, 357–61. Durand EY, Patterson N, Reich D, Slatkin M (2011) Testing for ancient admixture between closely related populations. Molecular biology and evolution, 28, 2239–52. Green RE, Krause J, Briggs AW et al. (2010) A draft sequence of the Neandertal genome. Science (New York, N.Y.), 328, 710–22. Hofreiter M (2001) DNA sequences from multiple amplifications reveal artifacts induced by cytosine deamination in ancient DNA. Nucleic Acids Research, 29, 4793–4799. Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England), 26, 589–95. Li H, Handsaker B, Wysoker A et al. (2009) The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England), 25, 2078–9. Li B, Zhang G, Willersleve E, Wang J (2011) GigaDB Dataset - DOI 10.5524/100008 Genomic data from the polar bear (Ursus maritimus). Miller W, Schuster SC, Welch AJ et al. (2012) Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proceedings of the National Academy of Sciences of the United States of America, 109, E2382– 90. 6