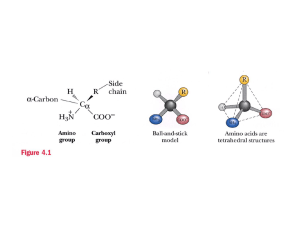
prot24315-sup-0001-suppinfo
advertisement

Supplementary information Crystal structure of At5g06450, a putative DnaQ-like exonuclease domain-containing protein from Arabidopsis thaliana, with novel homohexameric assembly Running title: Crystal structure of AtDECP David W. Smith,1* Mi Ra Han,2* Joon Sung Park,2 Kyung Rok Kim,2 Taeho Yeom,3 Ji Yeon Lee,2 Do Jin Kim,2 Craig A. Bingman,1 Hyun-Jung Kim,4 Kyubong Jo,3 Byung Woo Han,2† George N. Phillips, Jr.1,5 † 1 Center for Eukaryotic Structural Genomics, Department of Biochemistry, University of Wisconsin-Madison, Madison, Wisconsin 53706, USA 2 Research Institute of Pharmaceutical Sciences, College of Pharmacy, Seoul National University, Seoul 151-742, Korea 3 Department of Chemistry, Sogang University, Seoul 121-742, Korea 4 College of Pharmacy, Chung-Ang University, Seoul 156-756, Korea 5 Department of Biochemistry and Cell Biology, Rice University, Houston, TX 77251, USA * David W. Smith and Mi Ran Han contributed equally to this work. † Correspondence to: Byung Woo Han, College of Pharmacy, Seoul National University, Seoul 151-742, Korea. E-mail: bwhan@snu.ac.kr or George N. Phillips, Jr., Department of Biochemistry and Cell Biology, Rice University, Houston, Texas 77251, USA. E-mail: georgep@rice.edu 1 Size-exclusion chromatography profile analyses When we purified the His6-tagged AtDECP, we observed both hexameric and monomeric AtDECP fractions from the size-exclusion chromatography (HiLoadTM 16/600 Superdex200 pg, GE Healthcare) after Ni-NTA affinity chromatography. Two major fractions were eluted at the elution volume 67 mL and 123 mL. Calculated from the gel filtration protein standard (Bio-Rad), the fraction eluted at 67 mL corresponds to ~135 kDa, which is similar to the expected size of the hexameric AtDECP (~25 kDa/monomer x 6 = ~150 kDa). Even though the fraction eluted at 123 mL was calculated as much smaller protein than the AtDECP monomer, it was validated that the fraction contained the intact AtDECP monomer on SDSPAGE [Fig. S1]. Figure S1. Gel filtration chromatography profile and SDS-PAGE results of His6-tagged AtDECP after Ni-NTA affinity chromatography. The profile shows both hexameric and monomeric peaks of AtDECP. Red and blue inverted triangles represent the hexameric and monomeric AtDECP fractions, respectively. Interestingly, the monomeric AtDECP faction disappeared after the subsequent ion exchanged chromatography (HiTrap Q column, GE Healthcare) and most fractions were eluted as hexameric AtDECP fractions, which show the in vitro transition from monomer to hexamer AtDECP [Fig. S2]. 2 Figure S2. Gel filtration chromatography profile and SDS-PAGE results of the His6tagged monomeric AtDECP after the anion exchange chromatography. The profile shows a dominant hexameric AtDECP peak. Red inverted triangle represents the hexameric AtDECP fraction. In the case of the purification of the His6-maltose binding protein (MBP)-tagged AtDECP, we observed the only hexameric fraction from the size-exclusion chromatography after the removal of the His6-MBP tag using TEV proteases followed by second Ni-NTA affinity chromatography [Fig. S3] Figure S3. Gel filtration chromatography profile and SDS-PAGE results of the His6MBP-tagged AtDECP after the removal of His6-MBP tag using TEV proteases followed by second Ni-NTA affinity chromatography. The profile shows a dominant hexameric AtDECP peak. Red inverted triangle represents the hexameric AtDECP fraction. 3 Exonuclease activity assay of AtDECP To measure the exonuclease activity of AtDECP, we constructed and purified both His6MBP-tagged and His6-tagged AtDECP proteins. The purified AtDECP was co-incubated with oligonucleotides at 37 ℃ for 1 hour; AtDECP 21 μL and 33 μM 5’-overhang or gapped dsDNA 9 μL (final concentration of dsDNA = 10 μM). Positive control is 0.27 μM Klenow fragment (New England Biolabs) and is co-incubated with 10 μM dsDNA in the 1X reaction buffer (New England Biolabs) containing 50 mM NaCl, 10 mM Tris-HCl pH 7.9, 10 mM MgCl2, and 1 mM DTT at 37 ℃ for 30 min. The sequences of the co-incubated dsDNA are shown below. 5'-overhang dsDNA for Figure S4; 5'- GTACGACTGCAGGGA - 3’ 3'- CATGCTGACGTCCCTAGGAGCGCTTGTACCG -5' Gapped dsDNA by one nucleotide for Figure S5; 5'- GTACGACTGCAGGGA CCTCGCGAACATGGC-3' 3'- CATGCTGACGTCCCTAGGAGCGCTTGTACCG-5' Co-incubated samples were mixed with matrix solution onto Bruker MTP 384 target plate polished steel MALDI sample support. The matrix solution contains 350 mM 3hydroxypicolinic acid (Tokyo Chemical Industry, Tokyo, Japan), 80 mM pyrazinecarboxylic acid, and diammonium hydrogen citrate in 50% acetonitrile. MALDI-TOF MS analysis was performed with a Bruker AutoflaxTM speed machine (Bruker Daltonics Inc., Billerica, USA) in the linear positive ion mode using FlexControl 3.3 software for the automatic acquisition. 4 For the MALDI-TOF MS analysis with 3’-overhang dsDNA, AtDECP was co-incubated with oligonucleotides at 37 ℃ for 30 min; AtDECP 3 μL, 33 μM 3’-overhang DNA 9 μL (final concentration of dsDNA = 10 μM), and reaction buffer 18 μL containing 10 mM Tris-HCl pH7.9, 50 mM NaCl, 10 mM MgCl2, and 1 mM DTT. Positive control is T5 exonuclease (New England Biolabs) 1 unit in the same reaction condition. The sequences of co-incubated oligonucleotides are shown below. 3’-overhang dsDNA for Figure S6; 5' -CCTCGCGAACATGGC- 3' 3' -CATGCTGACGTCCCTGGAGCGCTTGTACCG- 5' The Klenow fragment showed 3’-5’ exonuclease activity with the presence of 5’-overhang and T5 exonuclease showed 5’-3’ exonuclease activity. However, in the case of AtDECP, the smaller fragment peaks, from the cleaved oligonucleotides due to the exonuclease activities, were not detected and the exonuclease activities of AtDECP could not be observed [Fig. S4S6]. 5 Figure S4. MALDI-TOF MS detection of cleaved DNA from 5’-overhang dsDNA. ‘*’ represents the shorter strand of dsDNA and ‘•’ represents the longer strand of dsDNA. (A) Hexameric AtDECP from the His6-MBP-tagged construct after the His6-MBP-tag removal (concentration: 3.129 mg/ml, buffer condition: 50 mM KH2PO4 pH 7.0, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, and 5% glycerol). (B) Monomeric AtDECP from the His6-tagged construct (concentration: 2.246 mg/ml, buffer condition: 10mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.1 mM DTT, and 5% Glycerol) (C) Hexameric AtDECP from the His6tagged construct (concentration: 0.24 mg/ml, buffer condition: 10mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.1 mM DTT, and 5% Glycerol). (D) Positive control, Klenow fragment (New England Biolabs). 6 Figure S5. MALDI-TOF MS detection of cleaved DNA from gapped dsDNA by one nucleotide. ‘*’ represents two shorter strands of dsDNA and ‘•’ represents the longer strand of dsDNA. (A) Hexameric AtDECP from the His6-MBP-tagged construct after the His6MBP-tag removal (concentration: 3.129 mg/ml, buffer condition: 50 mM KH2PO4 pH 7.0, 150 mM NaCl, 10 mM MgCl2, 5 mM DTT, and 5% glycerol). (B) Monomeric AtDECP from the His6-tagged construct (concentration: 2.246 mg/ml, buffer condition: 10mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.1 mM DTT, and 5% Glycerol) (C) Hexameric AtDECP from the His6-tagged construct (concentration: 0.24 mg/ml, buffer condition: 10mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.1 mM DTT, and 5% Glycerol). 7 Figure S6. MALDI-TOF MS detection of cleaved DNA from 3’-overhang dsDNA. ‘*’ represents the shorter strand of dsDNA and ‘•’ represents the longer strand of dsDNA. (A) Hexameric AtDECP from the His6-tagged construct after the ion-exchange chromatography (concentration: 0.148 mg/mL, buffer condition: 100 mM NaCl, 10 mM Tris-HCl pH 7.5, 0.1 mM DTT, and 5% glycerol) (B) Reconstituted hexameric AtDECP from the His6-tagged construct after the ion-exchange chromatography (concentration: 0.500 mg/mL, buffer condition: same as in A). (C) Hexameric AtDECP from the His6-tagged construct after the initial Ni-NTA affinity chromatography (concentration 0.594 mg/ml, buffer condition: same as in A). (D) Monomeric AtDECP from the His6-tagged construct after the initial Ni-NTA affinity chromatography (concentration: 9.932 mg/ml, buffer condition: same as in A). (E) Hexameric AtDECP from the His6-MBP-tagged construct after the His6-MBP-tag removal (concentration: 1.301 mg/ml, buffer condition: 150 mM NaCl and 50 mM KH2PO4, pH 7.0). (F) Positive control, 1 unit of T5 exonuclease (New England Biolabs). 8 dsDNA binding assay of AtDECP We implemented the EMSA to check if the purified AtDECP could bind with dsDNA in several different conditions. Three different types of dsDNA with 3’-overhang, 5’-overhang, and blunt-end were manufactured using oligo synthesis service (Cosmogenetech) and each overhang dsDNA contained five extra bases. Five AtDECP samples same as in Figure S6 were co-incubated with 3’-overhang dsDNA at 37 ℃ for one hour. Co-incubated samples were mixed with 6x DNA loading dye and electrophoretically eluted on 15% polyacrylamide gel. After the electrophoresis, the gel was visualized using TAE buffer with EtBr. We could not observe a gel shift of dsDNA from the interaction of AtDECP with dsDNA [Fig. S7]. The hexameric AtDECP sample, which was reconstituted from the His6-tagged construct after the ion-exchange chromatography, was co-incubated with three different types of dsDNA (5’-overhang, 3’- dsDNA, and blunt-ended) at 37 ℃ for one hour. Co-incubated samples were mixed with 6x DNA loading dye and electrophoretically eluted on 15% polyacrylamide gel. After the electrophoresis, the gel was visualized using TAE buffer with EtBr. We could not observe a gel shift of dsDNA from the interaction of AtDECP with dsDNA [Fig. S8]. 9 Lane 1 Lane 2 Lane 3 Negative control Mg2+ D.W. 10x buffer dsDNA AtDECP Total (μL) 28 2 30 + 25 3 2 30 Lane 4 Lane 5 A 27 2 1 30 Lane 6 Lane 7 B + 24 3 2 1 30 27 2 1 30 Lane 8 Lane 9 C + 24 3 2 1 30 27 2 1 30 Lane 10 Lane 11 D + 24 3 2 1 30 27 2 1 30 Lane 12 E + 24 3 2 1 30 27 2 1 30 + 24 3 2 1 30 Figure S7. Gel shift assay of dsDNA with AtDECP : (A) Hexameric AtDECP from the His6-tagged construct after the ion-exchange chromatography (concentration: 0.148 mg/mL, buffer condition: 100 mM NaCl, 10 mM Tris-HCl pH 7.5, 0.1 mM DTT, and 5% glycerol) (B) Reconstituted hexameric AtDECP from the His6-tagged construct after the ion-exchange chromatography (concentration: 0.500 mg/mL, buffer condition: same as in A). (C) Hexameric AtDECP from the His6-tagged construct after initial Ni-NTA (concentration 0.594 mg/ml, buffer condition: same as in A). (D) Monomeric AtDECP from the His6-tagged construct after initial Ni-NTA (concentration: 9.932 mg/ml, buffer condition: same as in A). (E) Hexameric AtDECP from the His6-MBP-tagged construct after the His6-MBP-tag removal (concentration: 1.301 mg/ml, buffer condition: 150 mM NaCl and 50 mM KH2PO4, pH 7.0). 10x buffer condition: 100 mM Tris, pH 7.5, 500 mM KCl, 10 mM DTT, 50 mM MgCl2. 10 Lane 1 Lane 2 Lane 3 Lane 4 Lane 5 Lane 6 Lane 7 Lane 8 Negative control Lane 9 Lane 10 Lane 11 Lane 12 Protein dsDNA/ Mn2+ 5’/ - 5’/ + 3’/ - 3’/ + Gap/ - Gap/ + 5’/ - 5’/ + 3’/ - 3’/ + Gap/ - Gap/ + DW 10x buffer dsDNA AtDECP 25 3 2 - 25 3 2 - 25 3 2 - 25 3 2 - 25 3 2 - 25 3 2 - 23 3 2 2 23 3 2 2 23 3 2 2 23 3 2 2 23 3 2 2 23 3 2 2 Total (μL) 30 30 30 30 30 30 30 30 30 30 30 30 Figure S8. Gel shift assay of three different types of dsDNA with the hexameric AtDECP. The protein used in this assay was the reconstituted hexameric AtDECP from the His6-tagged construct after the ion-exchange chromatography (concentration: 0.500 mg/mL, buffer condition: 100 mM NaCl, 10 mM Tris-HCl pH 7.5, 0.1 mM DTT, and 5% glycerol). 10x buffer condition without Mn2+: 100 mM Tris, pH 7.5, 500 mM KCl, 10 mM DTT. 10x buffer condition with Mn2+: 100 mM Tris, pH 7.5, 500 mM KCl, 10 mM DTT, 50 mM MnCl2. 11