Supplementary Methods (doc 52K)
advertisement

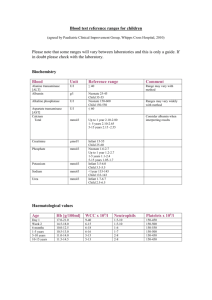
Supplementary Methods Acid substrates and RNAs All acid substrates were synthesized from the corresponding N-Boc protected amino acids (Watanabe chemical, Japan). All CME substrates were synthesized same procedure as previously described1. All DBE substrates were synthesized by coupling with 3,5-dinitrobenzylchloride (Sigma). All CBT substrates were synthesized by coupling with 4-chlrobenzyl mercaptan (TCI, Japan) using N,N-bis(2-oxo-3-oxazolidiyl)phosphorodiamidic chloride2. Proton (1H NMR) magnetic resonance spectra were recorded at 500 MHz on a Bluker AMX-500 spectrometer. The data are reported as follows: multipicity (s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad), coupling constant (Hz), integration. All oligonucleotides were purchased from Operon (Japan). The constructs of tRNAAsnXXX (XXX = CUA, ACCC, ACU, GUU or CUG) and microhelix RNA were synthesized using the same procedure as previously described3 (see also ref. 8 in main text). General procedure for synthesis of DBE substrates (valine 3,5-dinitrobenzyl ester): A mixture of -N-Boc-valine (130 mg, 0.60 mmol), triethylamine (101 mg, 1.0 mmol) and 3,5-dinitrobenzyl chloride (108 mg, 0.50 mmol) in 0.1 mL of dimethylformamide was stirred at room temperature for 12 h. After the reaction, diethylether (9 mL) was added and the solution was washed with 0.5 M HCl (3 mL x 3), 4 % NaHCO3 (3 mL x 3) and brine (5 mL x1), and the organic layer was dried over MgSO4 and concentrated under reduced pressure. The crude residue was dissolved in 2 mL of 4 M HCl/ethylacetate and incubated for 20 min at room temperarure. The solution 3 was concentrated under reduced pressue and the remained HCl was removed by repeating the addition of diethylether (3 mL) and concentration under reduced pressue three times. The product was precipitated by the addition of diethylether (3 mL) and the precipitants were filtered in 62 % overall yield (123 mg, 0.369 mmol).; 1H NMR (DMSO-d6, 500 MHz) 8.83 (s, 1H), 8.70 (s, 2H), 8.44 (br, 3H), 5.56 (s, 2H), 4.07 (d, J = 4.6 Hz, 1H), 2.22 (m, 1H), 1.00 (d, J = 7.0 Hz, 3H), 0.97 (d, J = 6.9 Hz, 3H). General procedure for synthesis of CBT substrates (valine 4-chlorobenzyl thioester): A mixture of -N-Boc-valine (108 mg, 0.50 mmol), triethylamine (152 mg, 1.5 mmol), N,N-bis(2-oxo-3-oxazolidiyl)phosphorodiamidic chloride (114 mg, 0.45 mmol) and 4-chlrobenzyl mercaptan (79 mg, 0.5 mmol) in 5 mL of dichloromethane is stirred at room temperature for 5 h. The solution was washed with 0.5 M HCl (3 mL x 3), 0.5 N NaOH (3 mL x 3) and brine (5 mL x1), and the organic layer was dried over MgSO4 and concentrated under reduced pressure. The crude residue was dissolved in 2 mL of 4 N HCl/ethylacetate and incubated for 20 min at room temperarure. The solution was concentrated under reduced pressue and the remained HCl was removed by repeating the addition of diethylether (3 mL) and concentration under reduced pressure three times. The product was precipitated by the addition of diethylether (3 mL) and the precipitants were filtered in 53 % overall yield (78.7 mg, 0.267 mmol); 1H NMR (DMSO-d6, 500 MHz) 8.33 (br, 3H), 7.40 (d, J = 8.5 Hz, 2H), 7.37 (d, J = 8.5 Hz, 2H), 4.27 (d, J = 14 Hz, 1H), 4.25 (d, J = 14 Hz, 1H), 4.19 (br, 1H), 2.17 (m, 1H), 0.94 (d, J = 6.9 Hz, 3H), 0.89 (d, J = 6.9 Hz, 3H). 4 dFx was synthesized by using the following procedure. P3 was annealed with dFxR46 (5'-ACCTA ACGCC ATGTA CCCTT TCGGG GATGC GGAAA TCTTT CGATC C-3'), and extended by Taq DNA polymerase. The resulting product was diluted 10 times with PCR reaction buffer and amplified by using P4 (5'-GCATA TGTAA TACGA CTCAC TATAG-3') and dFxR19 (5'-ACCTA ACGCC ATGTA CCCT-3') as the 5'- and 3'-primers, respectively. The DNA product was transcribed by T7 RNA polymerase and purified by 12% denaturing PAGE. eFx was synthesized by the same procedure as dFx except that eFxR45 (5'-ACCTA ACGCT AATCC CCTTT CGGGG CCGCG GAAAT CTTTC GATCC-3') and eFxR18 (5'-ACCT AACGC TAATC CCCT-3') was used instead of dFxR46 and dFxR19. The tRNA, dFx or eFx was independently dissolved in water and its concentration was adjusted to 200 µM. Acylation of tRNA and microhelix Reactions were carried out under the following conditions: 5 µL of 20 µM dFx or eFx, 20 µM tRNAAsnCUA, and 5 mM acid substrate in 0.1 M Hepes-K buffer pH 7.5, 0.1 M KCl, 600 mM MgCl2 and 20 % DMSO on ice. The procedure was as follows: 40 µM tRNAAsnCUA in 0.2 M Hepes-K buffer pH 7.5, 0.2 M KCl (2.5 µL) was heated at 95 ˚C for 3 min and cooled to 25 ˚C over 5 min. MgCl2 (3 M, 1 µL) and dFx or eFx (200 µM, 0.5 µL) were added and the mixture was stored at 25 ˚C for 5 min. The reaction was initiated by addition of 1 µL of 25 mM of the substrate in DMSO and incubated on ice for the listed times (Supplementary Table 1 online). 5 Analysis of acylation After the acylation of tRNAAsnCUA, the reaction was stopped by addition of 15 µL of 0.6 M sodium acetate pH 5 (with 5 mM DTT for Cys). The RNA was recovered by ethanol precipitation. The pellet was rinsed with 70 % ethanol containing 0.1 M NaCl, and then dissolved in 3.5 µL of 7.5 mg/mL sulfosuccinimidyl-D-biotin (Dojin, Japan) in 0.4 M Hepes-K pH 8.0. The biotinylation reaction was carried out on ice for 1 hour, and stopped by addition of 6.5 µL of 0.6 M sodium acetate pH 5 (with 5 mM DTT for Cys) followed by precipitation with ethanol. The pellet was rinsed twice with 70 % ethanol containing 0.1 M NaCl and dissolved in 100 µL of 10 mM sodium acetate pH 5 (with 5 mM DTT for Cys). 0.5 µL of this solution was mixed with 1.5 µL of loading buffer (0.2 mg/mL streptavidin in 37 mM piperazine pH 6.1, 37 mM EDTA and 6 M urea), and analyzed by 12 % denaturing PAGE (6 M urea). The RNA was stained with Syber Green II (Molecular Probe) and analyzed by FLA-5100 (Fuji, Japan). Acid PAGE analysis was carried out by the following procedure. After the acylation of microhelix RNA, the reaction was stopped by addition of 15 µL of 0.6 M sodium acetate pH 5 (with 5 mM DTT for Cys). Ethanol-precipitation followed by rinsing with 70% ethanol gave pellets which were then dissolved in 2.5 µL of 10 mM sodium acetate pH 5 (with 5 mM DTT for Cys). 1 µL of the solution was mixed with 1 µL of loading buffer (50 mM sodium acetate, 8 M urea) and analyzed by 20 % denaturing acid PAGE (50 mM sodium acetate, 6 M urea). The RNA was stained with ethidium bromide and analyzed by FLA-5100 (Fuji, Japan). 6 Calculation of suppression efficiency for protein site-specific mutagenesis The suppression efficiency was calculated based on the radioactivity of bands I and II, expressed as I/(I+IIx5/3); the intensity of band II was normalized in order to reflect the number of Met residues, based on the consideration that full-length and truncated proteins contain 5 and 3 Mets, respectively. We excluded band III from the calculation of suppression efficiencies, since this truncation might occur due to an abnormal termination, such as falling-off-termination of peptidyl-tRNAs from the ribosome. Preparation of mRNA for peptide synthesis Template DNA for peptide synthesis was prepared by the following procedure. (5'-GTAAT ACGAC TCACT ATAGG GTGAT CCAAC TTTAA TAAGG AGGTA TACCA ATG –3') was annealed with (5'-TCATC TTTAT AATCG GTGGT CTGCT GGTTG TTACT ACTCA TTGGT ATACC TCCTT ATT-3') and extended by Taq DNA polymerase. The resulting product was diluted 10 times with PCR reaction mixture and amplified using (5'-GTAATACGACTCACTATAGGG –3') and (5'- CGAAG CTTAT TTATC ATCAT CATCT TTATA ATCGG TGGT-3') as 5'- and 3'-primers, respectively. The DNA product was transcribed by T7 RNA polymerase and purified by 8 % denaturing PAGE. 7 Translation Acyl-tRNAAsnCUA was prepared by the following procedure. 40 µM of tRNAAsnCUA in 0.2 M Hepes-K buffer pH 7.5, 0.2 M KCl (12.5 µL) was heated at 95 ˚C for 3 min and cooled to 25 ˚C over 5 min. MgCl2 (3 M, 5 µL) and dFx or eFx (200 µM, 2.5 µL) were added and the solution was stored at 25 ˚C for 5 min. The reaction was initiated by addition of 5 µL of 25 mM substrate in DMSO and incubated on ice for the indicated times (Supplementary Table 1 online). The reaction was stopped by addition of 75 µL of 0.6 M sodium acetate pH 5 (with 5 mM DTT for Cys), and the RNA was recovered by ethanol precipitation. The pellet was rinsed twice with 70 % ethanol with 0.1 M sodium acetate pH 5.0, and once with 70 % ethanol. The pellet was dried and stored at –80 ˚C. The acyl-tRNA was dissolved in 3.5 µL of 1 mM sodium acetate and used for 3 rounds of translation. The batch translation (5 µL) was carried out using the rapid translation system, RTS-100® (Roche Diagnostics), in the presence of 35S-Met and analyzed by SDS-PAGE according to previous report (see ref. 9 in main text). Similarly, batch translation was performed using the PURE system (custom-made PURESYSTEM®, Post Genome Institute Co., ltd., Japan). The protein expression efficiency was determined based on our previous protocols and data published elsewhere9. The yields of the mutants under the optimal conditions was calculated by the equation of (suppression efficiency for each acid)/ (suppression efficiency for Iph) x 76 µg/mL. The suppression efficiency for each amino acid were shown in graphs Supplementary Figure 6a–d. We generally obtained the suppression efficiency of Iph to be 0.8, as seen in Supplementary Figure 6c, lane 31, a 76 µg/mL 8 expression efficiency was determined based on the quantification of Iph-GFP after the His-tag isolation. The translation of nonnatural peptides was carried out using the PURE system with 0.4 µM of mRNA and 3 mM EDTA. 200 µM of Met, Thr, Tyr, Lys, Ser, Asn, Gln and 50 µM of [14C]-Asp were added for natural peptide synthesis. 50 µM of each Aly-tRNAAsnACU, Cit-tRNAAsnGUU and Iph-tRNAAsnCUG were added instead of Ser, Asn and Gln for nonnatural peptide synthesis. The reaction was carried out in 2.5 µL at 37 ˚C for 1 hour and the products were analyzed by Tricine-SDS-PAGE. Mass spectra measurements of peptides For mass spectra analysis, the reactions (L) were performed in the absence of [14C]-Asp. The products were precipitated with 50 µL of acetone, dissolved in 2.5 µL of water, and then immobilized with FLAG-M2 agarose (Sigma). After the resin was washed with 50 µL of W buffer (50 mM Tris-HCl pH 8.0, 150 mM NaCl) twice, the immobilized peptides were eluted with 2.5 µL of 0.2 % TFA, desalted with C18 Zip tips (Millipore), and eluted with 1.5 µL of a 50% acetonitrile, 0.1% TFA solution saturated with the matrix (R)-cyano-4-hydroxycinnamic acid. Mass measurements were performed using MALDI-TOF (Autoflex®, BRUKER) in the positive mode and externally calibrated with Substance P (ave. 1,348.66 Da), Bombesin (ave. 1,620.88 Da), ACTH clip 1-17, (ave. 2,094.46 Da), Somatostatin 28, (ave. 3,149.61 Da) as standards. 9 References 1. 2. 3. Saito, H., Kourouklis, D. & Suga, H. EMBO J. 20, 1797-806 (2001). Diago-Meseguer, J., Palomo-Coll, A.L., Fernández-Lizarbe, J.R. & Zugaza-Bilbao, A. Synthesis 1980, 547-551 (1980). Kourouklis, D., Murakami, H. & Suga, H. Methods 36, 239-44 (2005). 10