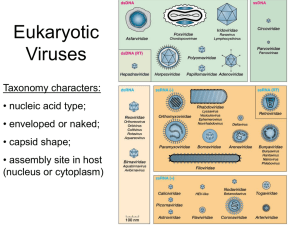
Classification of Viruses
advertisement

Classification of Viruses In this appendix selected groups of viruses are briefly described and a sketch of each is included. The illustrations are not to scale but should provide a general idea of the morphology of each group. The viruses are separated into five sections based primarily on their host preferences. This material has been adapted with permission from chapter 22 of Introduction to Modern Virology, 5th ed. by N. J. Dimmock, A. J. Easton, and K. N. Leppard, copyright © 2001 by Blackwell Scientific Publications, Ltd. We have not italicized the names of specific viruses in the text. However, it should be noted that in formal taxonomic usage the official name of a virus species—just as with other taxonomic names—is italicized, and the first letter of the first word in the species name is capitalized. Tentative species names and names of strains or isolates are not italicized. In this appendix, we will capitalize and italicize the genus name, but not always the species name. A. VIRUSES OF VERTEBRATE AND INVERTEBRATE ANIMALS 1. Viruses with double-stranded DNA genomes a. Family: Adenoviridae Nonenveloped icosahedral particles of 60 to 90 nm with a fiber protein at each vertex. Contains one molecule of linear double-stranded DNA (dsDNA) of 25,000 to 42,000 base-pairs (bp). A protein is covalently linked to the 5’ end and forms a pseudocircular genome by noncovalently linking to the 3’ end. Replicates and is assembled in the nucleus. Infects vertebrates but one genus infects fungi. Genera: Mastadenovirus—infects mammals (e.g., human adenovirus type 2 or type 5) Aviadenovirus—infects birds (e.g., fowl adenovirus type 1) Atendovirus—bovine adenoviruses Siadenovirus (e.g., frog adenovirus 1, turkey adenovirus 3) b. Family: Baculoviridae Enveloped rod-shaped particles, each with a nucleocapsid of 300 nm X 30 to 60 nm with one molecule of circular dsDNA of 80,000 to 180,000 bp. Virion may be occluded in a protein inclusion body (which makes it very stable). A large and diverse group, common in over 50 species of Lepidoptera; also infects Coleoptera, etc. Genera: Granulovirus—may be occluded in a polyhedron containing one particle (e.g., Cydia pomonella granulovirus). 1 Nucleopolyhedrovirus. Virions may be occluded in a polyhedron containing many particles (e.g., Autographica californica multiple nucleopolyhedrosis virus). c. Family: Herpesviridae Enveloped 120 to 220 nm particle with spikes, enclosing successively a tegument and an icosahedral nucleocapsid of 125 nm. One molecule of linear dsDNA of 120,000 to 240,000 bp. May bud from nuclear membrane. Very large family that infects vertebrates; latency is a common feature of the life cycle. Some are oncogenic. Subfamily: Alphaherpesvirinae Genera: Simplexvirus (e.g., human herpes [simplex] virus type 1 and 2—HHV-1 and -2) Varicellovirus (e.g., HHV-3 or varicella-zoster [chickenpox] virus) Subfamily: Betaherpesvirinae Genera: Cytomegalovirus (e.g., HHV-5 or human cytomegalovirus) Muromegalovirus (e.g., mouse cytomegalovirus) Roseolavirus (e.g., HHV-6, HHV-7) Subfamily: Gammaherpesvirinae (lymphoproliferative viruses; oncogenic) Genera: Lymphocryptovirus (e.g., HHV-4 or Epstein-Barr virus) Rhadinovirus (e.g., ateline herpesvirus 2, herpesvirus saimiri, HHV-8) Unclassified: Marek’s disease virus, turkey herpesvirus. d. Family: Iridoviridae Icosahedral isometric particle of 120 to 200 nm, which may be surrounded by a lipid envelope. Purified virus pellets are iridescent blue or green. One molecule of linear dsDNA of 140,000 to 300,000 bp, but virions may contain one or more copies. Contains several enzymes. Cytoplasmic assembly. Includes viruses of insects, fish (flounder, dab, and goldfish) and many frog species (e.g., frog virus 3). e. Family: Papillomaviridae Nonenveloped, 55 nm icosahedral particle with 72 capsomers in a skewed arrangement. One 8,000 bp molecule of double-stranded, covalently closed, circular DNA. Replicates and assembles in the nucleus. Over 60 human papillomaviruses (HPVs) and others from many species. Oncogenic. 2 f. Family: Polyomaviridae Nonenveloped 40 nm icosahedral particle with 72 capsomers in a skewed arrangement. One 5,000 bp molecule of covalently closed, circular dsDNA. Replicates and assembles in the nucleus. Includes murine polyomavirus and simian virus type 40 (SV40). Oncogenic. g. Family: Poxviridae Enveloped brick-shaped virions of 220 to 450 nm X 140 to 260 nm X 140 to 260 nm thick. Complex structure enclosing one or two lateral bodies and a biconcave core, with all enzymes required for mRNA synthesis. One molecule of linear ds-DNA of 130,000 to 375,000 bp. Cytoplasmic multiplication. Infects mostly vertebrates but also insects. Subfamily: Chondropoxvirinae (viruses of vertebrates) Genera: Orthopoxvirus (e.g. vaccinia and related viruses) Avipoxvirus (e.g., fowlpox and related viruses) Capripoxvirus (e.g., sheeppox and related viruses) Leporipoxvirus (e.g., myxoma of rabbits and related viruses; spread passively by arthropods) Molluscipoxvirus (e.g., molluscum contagiosum virus) Parapoxvirus (e.g., orf virus/milker’s node virus and related viruses) Suipoxvirus (e.g., swine pox virus) Yatapoxvirus (e.g., yaba/tanapox and related viruses of monkeys) Subfamily: Entomopoxvirinae (viruses of insects) Genera: Entomopoxvirus A, B, and C 2. Viruses with ssDNA genomes a. Family: Circoviridae Nonenveloped small icosahedral particles with circular ssDNA. Another genus infects plants. Genus: Circovirus—20 nm virion with one molecule of DNA of 1,800 to 2,300 nucleotides (e.g., chicken anemia virus; probably also TT [transfusion-transmitted] virus of humans) 3 b. Family: Parvoviridae Nonenveloped icosahedral particles of 18 to 26 nm that contain no enzymes. One 5,000 nucleotide linear molecule of either negative- or positive-sense ssDNA per particle. On extraction, these form an artifactual double strand. Replication is nuclear. Subfamily: Parvovirinae (viruses of vertebrates) Genera: Parvovirus—(e.g., canine and feline parvovirus, minute virus of mice) Erythrovirus—B19 virus, which causes fifth disease of children Dependovirus—these are satellite viruses Subfamily: Densovirinae (viruses of insects) 3. Viruses with dsRNA genomes and a virion-associated RNA-dependent RNA polymerase a. Family: Birnaviridae Nonenveloped icosahedral 60 nm particle with a 45 nm core containing two segments of linear dsRNA of 2,800 and 3,200 nucleotides. Has a VPg protein covalently linked to 5’ end of each segment. Cytoplasmic multiplication. Genera: Aquabirnavirus (e.g., pancreatic necrosis virus of fish) Avibirnavirus (e.g., infectious bursal disease of chickens, turkeys, ducks) Entomobirnavirus (e.g., Drosophila X virus) b. Family: Reoviridae Large family with members found in vertebrates, insects, and plants. Nonenveloped 60 to 80 nm icosahedral particle containing an isometric nucleocapsid with 10 to 12 segments of linear dsRNA of 18,000 to 30,000 bp. Cytoplasmic multiplication. Within a genus RNA segments in a mixed infection readily assort to form genetically stable hybrid virus. Genera: Orthoreovirus—infects vertebrates (humans, dogs, cattle, birds); 10 dsRNA segments (e.g., reoviruses of humans) Orbivirus—replicates in vertebrates and their insect vectors; 10 dsRNA segments (e.g., bluetongue virus of sheep) Rotavirus—name comes from its wheel-like particle with spokes; causes life-threatening diarrhea in very young vertebrates of many species, including humans; 11 dsRNA segments Aquaerovirus—viruses of fish and Crustacea; 11 dsRNA segments 4 Coltivirus—Colorado tick fever virus of vertebrates; 12 dsRNA segments Cypovirus—cytoplasmic polyhedrosis viruses of insects (including Lepidoptera, Hymenoptera, and Diptera); 10 dsRNA segments 4. Viruses with positive-sense ssRNA genomes a. Family: Caliciviridae Nonenveloped icosahedral 35 to 40 nm particle with 32 calyxlike (cup-shaped) depressions. Contains one molecule of linear positive-sense ssRNA of 7,500 nucleotides. The 5’ end has a VPg or a cap structure (hepatitis E virus). Cytoplasmic multiplication with subgenomic mRNA. Infects many species (e.g., human calicivirus, Norwalk virus, hepatitis E virus, rabbit hemorrhagic fever virus, swine exanthema virus). b. Family: Coronaviridae With the Arteriviridae, forms the order Nidovirales. Enveloped particles of 120 to 160 nm with club-shaped sparse protein spikes. Contains a helical nucleocapsid with one molecule of positivesense ss-RNA of 28,000 to 31,000 nucleotides. One of the largest RNA genomes. Cytoplasmic replication with a nested set of subgenomic mRNAs with a common leader sequence. Buds from Golgi apparatus and endoplasmic reticulum. Genera: Coronavirus (e.g., avian infectious bronchitis virus and human coronavirus OC43) Torovirus—biconcave or toroidal or doughnut-shaped particles (Berne virus of horses and Breda virus of cattle) c. Family: Flaviviridae Enveloped 40 to 60 nm particles with an isometric nucleocapsid of 25 to 30 nm and one molecule of linear positive-sense ss-RNA of 9,500 to 12,500 nucleotides. Differ from Alphaviridae by the presence of a matrix protein, the lack of subgenomic mRNAs, and budding from the endoplasmic reticulum. Cytoplasmic multiplication. Spread by arthropods unless stated. Genera: Flavivirus—large group of viruses that multiply in the vertebrate host and insect or tick vector (e.g., yellow fever virus, tick-borne encephalitis virus group, dengue virus group, Japanese encephalitis virus group) Pestivirus—includes border disease virus (sheep), bovine diarrhea virus, classic swine fever (hog cholera) virus; no vector 5 Hepacivirus—hepatitis C virus of humans; no vector d. Family: Picornaviridae Nonenveloped viruses of mainly vertebrates. One molecule of linear positive-sense ssRNA of 7,000 to 8,500 nucleotides in 30 nm icosahedral particles. Has a 5’ VPg protein. Cytoplasmic multiplication. Genera: Enterovirus—acid resistant, primarily of the intestinal tract (e.g., polioviruses, most echoviruses, coxsackieviruses of humans and various nonhuman enteroviruses) Aphthovirus—economically important foot-and-mouth disease viruses of cattle and other ruminants Cardiovirus (e.g., encephalomyocarditis [EMC] virus of mice) Hepatovirus (e.g., human and simian hepatitis A viruses) Parechovirus (e.g., human echoviruses 22 and 23) Rhinovirus—acid labile; infects the upper respiratory tract; includes about 100 common cold viruses; also equine rhinoviruses and various unclassified viruses of insects e. Family: Tetraviridae Nonenveloped 40 nm icosahedral particle with one molecule (6,500 nucleotides) or two molecules (2,500 and 5,300 nucleotides) of linear positive-sense ssRNA. No infection of cultured cells. All isolated from Lepidoptera (e.g., Naudaurelia virus) f. Family: Togaviridae Enveloped 70 nm particles containing an icosahedral nucleocapsid and one molecule of linear positive-sense ssRNA of 9,000 to 12,000 nucleotides. Cytoplasmic multiplication; buds from the plasma membrane. Has an intracellular subgenomic mRNA. Genera: Alphavirus—multiplies in vertebrate host and insect vector (e.g., Sindbis virus and Sernliki forest virus) Rubivirus—rubella virus of humans; no vector 6 5. Viruses with negative-sense/ambisense ssRNA genomes and a virion-associated RNAdependent RNA polymerase a. Family: Arenaviridae Enveloped isometric usually 100 nm particles with club-shaped spikes. Genome is contained in two circular helical nucleocapsids, the larger with one molecule of linear negative-sense ssRNA of 7,500 nucleotides and the smaller, also linear, of 3,500 nucleotides. Both are ambisense. May package more than two nucleocapsids per virion. Virion contains host cell ribosomes for no known function. Cytoplasmic multiplication; buds from plasma membrane. Divided into the LCM/LASV subgroup (Old World arenaviruses: lymphocytic choriomeningitis virus, lassa virus and related viruses) and the Tacaribe complex subgroup (New World arenaviruses: Tacaribe, Junin, Pichinde and related viruses). b. Family: Bornaviridae Enveloped isometric 90 nm virions with a helical nucleocapsid and one molecule of linear negative-sense ssRNA of 9,000 nucleotides. Nuclear replication. Recently isolated from people with behavioral/neuropsychiatric problems but not known if causal. Known to infect horses since the eighteenth century and several other vertebrate species. Natural host unknown. Borna disease virus. c. Family: Bunyaviridae Enveloped isometric 100 nm particle with 10 nm spikes. Contains three helical nucleocapsids, each with one molecule of linear negative-sense ssRNA—large: 6,000 to 12,000 nucleotides, medium: 3,500 to 6,000 nucleotides, and small: 1,000 to 2,000 nucleotides. Cytoplasmic multiplication; buds from the Golgi membranes. Infects vertebrates and spread by arthropods unless stated. Also a genus that infects plants. Genera: Bunyavirus—mosquito and gnat vectors (e.g., Bunyamwera and 150 or so other viruses) Hantavirus—no arthropod vector (e.g., Hantaan virus) Nairovirus—tick vector (e.g., Nairobi sheep disease virus) Phlebovirus—S RNA is ambisense; sandfly, tick and gnat vectors (e.g., sandfly fever virus, uukuniemi virus) d. Family: Filoviridae 7 Genus: Filovirus—enveloped long filamentous, sometimes branched, particles 800 to 900 (sometimes 14,000) nm X 80 nm with helical nucleocapsid of 50 nm diameter. One molecule of negativesense ssRNA of 19,000 nucleotides. Buds from plasma membrane. Includes Marburg, Ebola, and Reston viruses. Highly pathogenic for humans. Natural reservoir not known. e. Family: Orthomyxoviridae Enveloped pleomorphic 100 nm (sometimes filamentous) particles with a dense layer of protein spikes. Six to eight helical nucleocapsids, 9 nm in diameter with transcriptase activity. Each contains one molecule of linear negative-sense ssRNA, totaling 12,000 to 15,000 nucleotides. All RNA synthesis is nuclear. Within a genus, RNA segments in a mixed infection readily assort to form genetically stable hybrid viruses. Plasma membrane budding. Genera: Influenzavirus A—genome comprises eight molecules of RNA ranging from 900 to 2,350 nucleotides. Virions have separate hemagglutinin and neuraminidase spike proteins. Undergoes antigenic shift and drift. Natural reservoir is seashore birds; infects several vertebrate species including humans. Influenzavirus B—genome comprises eight molecules of RNA ranging from 900 to 2,350 nucleotides. Virions have separate hemagglutinin and neuraminidase spike proteins. Undergoes antigenic drift only. Infects humans. Influenzavirus C—genome comprises seven molecules of RNA ranging from 1,000 to 2,350 nucleotides. Virions have one hemagglutinin–esterase fusion spike protein. The esterase is a receptor-destroying enzyme. Undergoes minor antigenic variation. Infects humans. Thogotovirus—carried by ticks and occasionally infects humans. Genome comprises six molecules of RNA. (Thogoto virus and Dhori virus; Asia, Africa, and Europe.) Possibly also another genus for infectious salmon anemia virus. f. Family: Paramyxoviridae Enveloped pleomorphic particle usually 150 to 200 nm in diameter. Has a dense layer of fusion protein spikes and attachment protein spikes. Contains one helical nucleocapsid 13 to 18 nm in diameter with one molecule of linear negative-sense ss-RNA of 13,000 to 16,000 nucleotides. Filamentous forms of up to 400 nm common. Cytoplasmic multiplication; buds from plasma membrane. Infects vertebrates. Most, but not all, are respiratory viruses. Subfamily: Paramyxovirinae 8 Genera: Respirovirus (e.g., human parainfluenzavirus type 1) Morbillivirus—measles virus, rinderpest virus, and canine distemper virus Rubulavirus (e.g., mumps virus, Newcastle disease virus of poultry or avian parainfluenzavirus type 1) Subfamily: Pneumovirinae Genera: Pneumovirus (e.g., human and bovine respiratory syncytial viruses, pneumonia virus of mice) Metapneumovirus (e.g., turkey rhinotracheitis virus) g. Family: Rhabdoviridae Enveloped, bullet-shaped particles 100 to 430 nm X 45 to 100 nm, containing one molecule of linear negative-sense ssRNA of 11,000 to 15,000 nucleotides. Has 5–10 nm spikes. Inside is a helical nucleocapsid. Cytoplasmic multiplication and buds from plasma membrane. Infects vertebrates. Also genera that infect plants. Genera: Vesiculovirus (e.g., vesicular stomatitis virus) Lyssavirus (e.g., rabies virus, unusual as infects all mammals) Ephemerovirus (e.g., bovine ephemeral fever virus) Novirhabdovirus (e.g., infectious hemopoietic virus of fish) 6. Viruses with RNA genomes that replicate through a DNA intermediate a. Family: Retroviridae Enveloped 80 to 100 nm particles with spikes. Nucleocapsid can be isometric or a truncated cone; contains two identical copies of a linear positive-sense ssRNA of 7,000 to 11,000 nucleotides. Virions contain reverse transcriptase and integrase enzymes. The DNA provirus is nuclear and integrated with host DNA. Transmission is horizontal or vertical. Associated with many different diseases. Not all viruses are oncogenic. Genera: “Mammalian type C retroviruses” (e.g., murine leukemia virus); oncogenic Alpharetrovirus (e.g., avian leukosis virus, Rous sarcoma virus); oncogenic Betaretrovirus (e.g., Mason-Pfizer monkey virus) Gammaretrovirus (e.g., mouse mammary tumour virus); oncogenic Deltaretrovirus (e.g., bovine leukemia virus, human T-cell lymphotrophic virus types 1–3); oncogenic Epsilonretrovirus (e.g., walleye dermal sarcoma virus of fish) 9 Lentivirus—infects primates (human immunodeficiency virus types 1 and 2, simian immunodeficiency viruses), horses, sheep, goats, cattle, and cats; all associated with immunodeficiency diseases Spumavirus (e.g., human spumaviruses); named after their foamy cytopathogenic effect; no disease known 7. Viruses with a DNA genome that replicate through an RNA intermediate a. Family: Hepadnaviridae A40 to 48 nm enveloped particle containing an isometric nucleocapsid with DNA polymerase and protein kinase activities. One partially double-stranded circular DNA molecule that is not covalently closed. This has a complete negative-sense strand of 3,000 nucleotides with a 5’terminal protein, and a variable length positive-sense strand of 1,700 to 2,800 nucleotides. The circularization overlaps the 3’ and 5’ termini of the negative-sense DNA. These are “reversiviruses” that have a reverse transcriptase. Genera: Orthohepadnavirus (e.g., hepatitis B [HBV] of humans, which is strongly associated with liver cancer, and woodchuck hepatitis virus) Avihepadnavirus (e.g., duck hepatitis B virus) B. VIRUSES THAT MULTIPLY IN PLANTS Knowledge of plant virus multiplication is harder to obtain than that of animal viruses because plant cell culture systems are less manageable. Greater emphasis is given to physical properties and disease characteristics, especially as many are important in agriculture. Differences in virus proteins, translation strategy (which may not be mentioned here), and vector are important criteria in plant virus classification. Some plant viruses also multiply in their animal vector (Bunyaviridae, Marafivirus, Reoviridae, Rhabdoviridae), so the plant/animal virus distinction becomes uncertain. 1. Viruses with ssDNA genomes a. Family: Geminiviridae Virions comprise two incomplete icosahedra, 30 nm long X 18 nm, joined as Siamese twins. Nonenveloped. Has closed circular ssDNA. Nuclear replication. Persistent or not in an insect vector but does not multiply in the vector. Genera: Curtovirus—genome is one DNA molecule of 3,000 nucleotides. Does not infect monocotyledonous plants (grasses). Whitefly vector (e.g., beet curly top virus). Mastrevirus—genome is one DNA molecule of 2,600 nucleotides. Infects mainly monocotyledonous plants (grasses). Leafhopper vector (e.g., maize streak virus). Begomovirus—most genomes are two molecules of DNA each of 2,500 nucleotides. Whitefly vector (e.g., bean golden mosaic virus). 10 2. Viruses with dsRNA genomes and a virion-associated RNA-dependent RNA polymerase a. Family: Reoviridae Large and widespread family. Genera also found in vertebrates and insects. Nonenveloped 60 to 80 nm icosahedral particles containing an isometric nucleocapsid with 10 to 12 segments of linear dsRNA of 18,000 to 30,000 bp. Has a transcriptase activity. Cytoplasmic multiplication. Within a genus RNA segments in a mixed infection readily assort to form genetically stable hybrid virus. Genera: Fijivirus—10 dsRNA segments. Multiplies in leafhopper insect vector (e.g., Fiji disease virus). Oryzavirus—10 dsRNA segments. Multiplies in insect vector (e.g., rice ragged stunt virus). Phytoreovirus—12 dsRNA segments. Multiplies in leafhopper insect vector (e.g., wound tumour virus of clover). 3. Viruses with positive-sense ssRNA genomes Isometric virions a. Family: Comoviridae Two molecules of linear positive-sense ssRNA, each encapsidated separately in a nonenveloped 30 nm icosahedral particle. Both RNAs needed for infectivity. Cytoplasmic multiplication. Genera: Comovirus—RNAs of < 7,000 and 3,981 nucleotides. Nonpersistent in beetle vector (e.g., cowpea mosaic virus). Fabavirus—RNAs of < 7,000 and 4,500 nucleotides. Nonpersistent in aphid vector (e.g., broad-bean wilt virus 1). Nepovirus—RNAs of > 7,000 and 4,000 nucleotides. Does not multiply in nematode vector or not vectored (e.g., tobacco ringspot virus). b. Family: Luteoviridae Nonenveloped icosahedral 25 to 30 nm particles with one molecule of linear positive-sense ssRNA of 5,500 to 7,000 nucleotides. Does not multiply in aphid vector, but persists. Genera: Enamovirus—no VPg protein (e.g., pea enation mosaic virus) Luteovirus—no VPg (e.g., barley yellow dwarf virus) 11 Polerovirus—RNA has a 5’ VPg (e.g., potato leafroll virus) c. Family: Tombusviridae Nonenveloped isometric 32 to 35 nm particle with one or two molecules of linear positive-sense ssRNA totaling 4,000 to 5,000 nucleotides. Cytoplasmic multiplication. Genera: Aureusvirus—one RNA. Soil transmitted; no vector (e.g., Pothos latent virus). Avenavirus—one RNA. Transmitted mechanically from plant to plant and possibly also by fungi (e.g., oat chlorotic stunt virus). Carmovirus—one RNA. Soilborne transmission without vector (e.g., carnation mottle virus). Dianthovirus—two RNAs (4,000 and 1,500 nucleotides) in one particle. Soilborne transmission without vector (e.g., carnation ringspot virus). Machlomovirus—one RNA. Infects only grasses. Mechanical or beetle transmission (e.g., maize chlorotic mottle virus). Necrovirus—one RNA. Fungal vector (e.g., tobacco necrosis virus A). Panicovirus—one RNA. Transmitted mechanically (e.g., Panicum mosaic virus). Tombusvirus—one RNA. Transmitted mechanically or by grafting (e.g., tomato bushy stunt virus). Isometric virions and virions that are short rods d. Family: Bromoviridae Nonenveloped particles. Linear positive-sense ssRNA. Tripartite genome of 2,000 to 3,000 nucleotides (total 8,000 nucleotides). RNA 4 is coat protein mRNA encoded by RNA 3. Cytoplasmic multiplication. Genera: Alfamovirus—four RNAs in four bacilliform particles (short rods) 56, 43, 35, and 30 nm X 18 nm. Particles each with one molecule of RNA 1 (3,644 nucleotides) or RNA 2 (2,593 nucleotides) or RNA 3 (2,037 nucleotides) or two molecules of RNA 4 (881 nucleotides). Infectivity requires RNAs 1 to 3 + coat protein or RNA 4. Cytoplasmic multiplication. Nonpersistent in aphid vector (e.g., alfalfa mosaic virus). 12 Bromovirus—four RNAs in three isometric 27 nm particles. Particles each with one molecule of RNA 1 (3,234 nucleotides) or RNA 2 (2,865 nucleotides) or RNA 3 (2,117 nucleotides) + RNA 4 (800 nucleotides). Infectivity requires RNAs 1 to 3 + coat protein or RNA 4. Beetle vectors (e.g., brome mosaic virus). Virions that are rigid rods These genera have not yet been assigned to a family: e. Tobamovirus—Nonenveloped rod, 300 nm X 18 nm, with helical symmetry. Contains one molecule of linear positive-sense ssRNA of 6,395 nucleotides. Transmitted mechanically or by seed (e.g., tobacco mosaic virus). f. Tobravirus—two nonenveloped rods, 200 nm X 22 nm and 46 to 115 nm X 22 nm, with helical symmetry. Contains, respectively, one molecule of linear positive-sense ssRNA of 7,000 or 2,000 to 4,000 nucleotides. The larger RNA alone is infectious and the smaller one specifies the coat protein. Both are needed for synthesis of new virions. Cytoplasmic multiplication. Nematode vector (e.g., tobacco rattle virus). Virions that are flexuous rods (i.e., with bends but not necessarily flexible) g. Family: Potyviridae Nonenveloped flexuous rod(s) with helical symmetry. There are one or two molecules of linear positive-sense ss-RNA. Cytoplasmic multiplication. Genera have different vectors. Genera: Potyvirus—particle of 700 to 900 nm X 12 nm with one RNA molecule of 10,000 nucleotides. A VPg at the 5’ end. Nonpersistent in aphid vector (e.g., potato virus Y). Ipomovirus—one particle of 800 to 950 nm X 12 to 16 nm with one RNA molecule of 11,000 nucleotides. Whitefly vector (e.g., sweet potato mild mottle virus). 13 Macluravirus—nonenveloped flexuous rod, 700 nm X 13 to 16 nm, with one molecule of RNA of 8,000 nucleotides. Nonpersistent in aphid vector (e.g., maclura mosaic virus). Rymovirus—one particle of 700 nm X 11 to 15 nm with one molecule of RNA of 8,500 to 10,000 nucleotides. Mite vector (e.g., ryegrass mosaic virus). Tritimovirus—one particle of 700 nm _ 13 nm with one molecule of RNA of 9,672 nucleotides. Persistent in mite vector (e.g., wheat streak mosaic virus). Bymovirus—two particles of 500 to 600 and 250 to 300 nm X 13 nm. The larger with an RNA of 7,632 nucleotides and the smaller with an RNA of 3,585 nucleotides. Fungal vector (e.g., barley yellow mosaic virus). 4. Viruses with negative-sense/ambisense ssRNA genomes and a virion-associated RNAdependent RNA polymerase a. Family: Bunyaviridae Enveloped isometric 100 nm particle with 10 nm spikes. Each contains three helical nucleocapsids with linear negative-sense ssRNA. The largest nucleocapsid comprises an RNA of 6,000 to 12,000 nucleotides, the medium of 3,500 to 6,000 nucleotides, and the smallest of 1,000 to 2,000 nucleotides. Cytoplasmic multiplication; buds from the Golgi membranes. Spread by arthropods unless stated. Also genera that infect animals. Genus: Tospovirus—M and S RNAs are ambisense. Transmitted by insect (thrips) vector. Multiplies in vector (e.g., tomato spotted wilt virus). b. Family: Rhabdoviridae Enveloped, bullet-shaped particles 100 to 430 nm long and 45 to 100 nm in diameter, containing a helical nucleocapsid with one molecule of linear negative-sense ssRNA of 11,000 to 15,000 nucleotides. Has 5 to 10 nm spikes. Also genera that infect animals. Genera: Cytorhabdovirus—plant viruses that mature in the cytoplasm. Spread by (and multiply in) insect vector (e.g., lettuce necrotic yellows virus). Nucleorhabdovirus—plant viruses that mature in the nucleus. Spread by (and multiply in) insect vector (e.g., potato yellow dwarf virus). 14 5. Viruses with DNA genomes that replicate through an RNA intermediate a. Family: Caulimoviridae A nonenveloped particle containing one partially double-stranded circular DNA molecule that is not covalently closed (like that of hepadnaviruses), with a complete negative-sense strand of 7,000 to 8,000 nucleotides and an incomplete positive-sense strand with one to three discontinuities. The circularization, overlaps the 3’ and 5’ termini of the negative-sense DNA. These are “reversiviruses” that have a reverse transcriptase. Replication is nuclear and the genome remains episomal and does not integrate. Genera: Caulimovirus—icosahedral 50 nm particles. Nonpersistent in aphid vectors (e.g., cauliflower mosaic virus). Badnavirus—rod-shaped particles 130 nm X 30 nm. Nonpersistent in beetle larvae (e.g., commelina yellow mottle virus). C. VIRUSES MULTIPLYING IN ALGAE, FUNGI, AND PROTOZOA 1. Viruses with dsDNA genomes a. Family: Adenoviridae Nonenveloped icosahedral particles of 60 to 90 nm with a fiber protein from each vertex. Contains one molecule of linear ds-DNA of 25,000 to 42,000 bp. Replicate and assembled in the nucleus. Other genera infect animals. (Putative) genus: Rhizidiovirus—60 nm virion with one molecule of linear dsDNA of 25,000 bp. Infects fungi (e.g., Rhizidiomyces virus). b. Family: Phycodnaviridae Infects algae. Nonenveloped, isometric, 130 to 190 nm, multilayered particles containing one molecule of linear dsDNA of 160,000 to 380,000 bp. Contain internal lipid. Infects Paramecium, Chlorella, and Hydra spp. May have a use in controlling algal blooms. 15 2. Viruses with dsRNA genomes and a virion-associated RNA-dependent RNA polymerase a. Family: Hypoviridae Genus: Hypovirus—vesicles of 50 to 80 nm with one molecule of linear dsRNA of 9,000 to 13,000 bp, no structural proteins and polymerase activity. Surrounded by rough endoplasmic reticulum in infected cells. Causes hypovirulence of host chestnut blight fungus (e.g., Cryphonectria hypovirus 1). b. Family: Partitiviridae Two molecules of linear dsRNA of 1,400 to 3,000 bp packaged in one or several nonenveloped isometric particles that lack structural detail. Have a transcriptase. Cytoplasmic multiplication. Also genera that infect plants. Genera: Partitivirus—particles 30 to 35 nm. Infects fungi, (e.g., Gaeumannomyces graminis virus 019/6A). Chrysovirus—particles 35 to 40 nm. Infects fungi (e.g., Penicillium chrysogenum virus). c. Family: Totiviridae Nonenveloped particles of 30 to 40 nm with one molecule of linear dsRNA of 5,000 to 7,000 bp. One major capsid protein. Single shell. Cytoplasmic multiplication. Genera: Totivirus—some viruses also have other satellite RNA segments that encode killer proteins. Infects the fungus Saccharomyces cerevisiae. Giardiavirus—infects the protozoan Giardia lamblia. Leishmaniavirus—one RNA. Infects protozoa: Leishmania spp. 3. Viruses with positive-sense ssRNA genomes a. Family: Barnaviridae Nonenveloped icosahedral short rod, 50 nm X 19 nm, with one molecule of linear ssRNA of 4,009 nucleotides. Infects fungi (e.g., mushroom bacilliform virus). 16 b. Family: Narnaviridae One molecule of linear positive-sense ssRNA of 2,500 nucleotides as a ribonucleoprotein with polymerase activity. Infects fungi. Genera: Narnavirus (e.g., Saccharomyces cerevisiae 20S narnavirus) Mitovirus (e.g., Cryphonectria parasitica mitovirus 1 NB631) D. VIRUSES (PHAGES) MULTIPLYING IN ARCHAEA, BACTERIA, MYCOPLASMA, AND SPIROPLASMA The well-known molecular biology of phages is based on a detailed study of a few representatives, and surprisingly little is known of their comparative biology. 1. Viruses with dsDNA genomes Viruses that have some head-tail structure, here ordered by decreasing tail length a. Family: Siphoviridae Nonenveloped particle with a long noncontractile tail up to 570 nm X 8 nm. Icosahedral head of 60 nm. Contains one molecule of linear dsDNA of 48,500 bp. Causes no host DNA breakdown. Infects bacteria including enterobacteria, mycobacteria, Lactococcus, Methanobacterium, and Vibrio. Includes coliphage T1, lambda (λ), chi (), and phi () 80. b. Family: Myoviridae Nonenveloped particle with a complex contractile tail 80 to 455 nm X 16 nm. Contraction requires ATP. Head separated from tail by a neck. Head isometric (as shown) or elongated, 110 nm X 80 nm. Has one molecule of linear dsDNA of 40,000 to 170,000 bp. Infects enterobacteria, Bacillus, and Halobacterium spp. (Archaea). Includes the T-even coliphages T2, T4, and T6, and PBS1, SP8, SP50, P1, P2, 21, 34, and Mu. 17 c. Family: Podoviridae Nonenveloped particle with a short noncontractile tail of 20 nm X 8 nm. Icosahedral head of 60 nm. One molecule of linear dsDNA of 19,000 to 44,000 bp. Causes host DNA breakdown. Includes the coliphages T3 and T7, enterobacteria phage P22, and bacillus phage phi () 29. Viruses that do not have a head-tail structure d. Family: Fuselloviridae Enveloped lemon-shaped particle 100 nm X 60 nm with short tail fibers at one pole. One molecule of circular dsDNA of 15,465 bp. Infects Archaea such as Desulfolobus and Methanococcus (e.g. Sulfolobus virus 1). e. Family: Tectiviridae Nonenveloped icosahedral 63 nm particle. Unusual enveloped nucleoprotein surrounded by lipid. One molecule of linear dsDNA of 150,000 bp. On attachment forms a tail-like tube 60 nm X 10 nm. Infects gram-negative bacteria (e.g., enterobacteria phage PRD1). Has similarities with adenoviruses. f. Family: Corticoviridae Nonenveloped, nontailed icosahedral 60 nm particle formed of several layers, including one of lipid. Spikes at vertices. One molecule of circular dsDNA of 9,000 bp. Infects Pseudomonas (e.g., Alteromonas phage PM2). g. Family: Plasmaviridae 18 Enveloped pleomorphic 80 nm particle with small dense core; 50 and 125 nm particles also produced. One molecule of circular dsDNA of 12,000 bp. Infects Mycoplasma (e.g., Acholeplasma phage L2). h. Family: Rudiviridae Nonenveloped rigid rod of 500 to 780 nm X 23 nm with helical symmetry, with a plug and three tail fibers at each end. One molecule of linear dsDNA of 33,000 bp. Nonlytic. Infects thermophilic Archaea, Sulfolobus (e.g., Sulfolobus virus SIRV-1). i. Family: Lipothrixviridae Enveloped, rigid rod of 400 nm X 40 nm with protrusions at both ends that participate in cell attachment. One molecule of linear dsDNA of 16,000 bp. Virions stable at 100°C. Infects Archaea (e.g., Thermoproteus virus 1). 2. Viruses with ssDNA genomes a. Family: Inoviridae Nonenveloped rod with one molecule of circular positive-sense ssDNA of 5,000 to 10,000 nucleotides. Virion length depends on the length of the DNA and its conformation. Genera: Inovirus—flexuous rods of 700 to 2,000 nm X 7 nm with DNA of 4,400 to 8,500 nucleotides. Host bacteria not lysed. Includes enterobacteria phage M13 and enterobacteria phage fd. Plectrovirus—nearly straight rods of 85 or 280 nm X 10 to 15 nm with DNA of 4,400 to 8,500 nucleotides. Infects Mycoplasma (85 nm) and Spiroplasma (280 nm) (e.g. Acholeplasma phage MV-L51). 19 b. Family: Microviridae Icosahedral particle with large knobs on the 12 vertices. Minimum and maximum diameters are 22 and 23 nm, respectively. One molecule of circular ssDNA of 4,400 to 5,400 nucleotides. Genera: Microvirus—infects bacteria (e.g., enterobacteria phage phi () X 174) Spiromicrovirus—infects Spiroplasma (e.g., Spiroplasma phage 4) Chlamydiamicrovirus (e.g., Chlamydia phage 1) Bdellomicrovirus (e.g., Bdellovibrio phage MAC 1) 3. Viruses with dsRNA genomes and a virion-associated RNA-dependent RNA polymerase a. Family: Cystoviridae Enveloped icosahedral 85 nm particle with 8 nm spikes. Has a 58 nm nucleocapsid and a 43 nm core. Each particle contains three molecules of linear dsRNA of 6,374, 4,057, and 2,948 bp. Infects Pseudomonas (e.g., Pseudomonas phage phi [] 6). 4. Viruses with positive-sense ssRNA genomes a. Family: Leviviridae Nonenveloped 26 nm icosahedral particles with one molecule of linear positive-sense ssRNA of 3,400 to 4,300 nucleotides. Include enterobacteria phages R17, MS2, and Q. E. SATELLITE VIRUSES AND SATELLITE NUCLEIC ACIDS OF ANIMALS, PLANTS, FUNGI, AND BACTERIA The replication of satellite viruses and satellite nucleic acids depends upon coinfection of a host cell with a helper virus. The satellite genome has no significant homology with that of the helper and hence differs from other types of dependent nucleic acid molecules. Here, we use a broad definition of a satellite virus—one that is incapable of independent production of progeny virus particles, yet may be able to replicate itself. Satellite viruses encode their own coat protein, whereas satellite nucleic acids do not and use the coat protein of their helper virus. Some satellite viruses/satellite nucleic acids modulate the replication of the helper virus and exacerbate or diminish disease. 20 1. Satellite nucleic acids with dsDNA genomes Nonenveloped isometric virions with a Myoviridae helper (enterobacteria phage P2). One molecule of linear dsDNA of 11,627 bp. Interesting that the head-tail helper virion protein can form an isometric virion (e.g., enterobacteria P4 satellite). 2. Satellite viruses with ssDNA genomes a. Family: Parvoviridae Nonenveloped icosahedral particle of 18 to 26 nm that contains no enzyme. One 5,000 nucleotide linear molecule of either negative- or positive-sense ssDNA per particle. On extraction these form an artifactual double strand. Nuclear replication. There are also independently replicating (autonomous) parvoviruses of animals. Subfamily: Parvovirinae Genus: Dependovirus—dependent on helper adenovirus or herpesvirus for efficient replication, but in some cell cultures replicate independently. Particles package 90% negative-sense DNA. Encode their own coat protein. Adeno-associated viruses. Infects vertebrates. 3. Satellite nucleic acids with ssDNA genomes A genome of 682 nucleotides with no ORF. Geminiviruses helpers (e.g., tomato leaf curl virus satellite DNA). Infects plants. Particle morphology not yet determined. 4. Satellite nucleic acids with dsRNA genomes Nonenveloped isometric particles with a Totiviridae helper virus. Linear dsRNA of 500 to 1,800 bp inside a virion made of helper virus coat protein (e.g., satellite of Saccharomyces cerevisiae M virus). Infects fungi. 5. Satellite viruses with positive-sense ssRNA genomes Isometric particles with one linear ssRNA. Tobacco necrosis virus satellite virus subgroup: Particles of 17 nm containing one molecule of RNA of 1,239 nucleotides. Fungal vector. Infects plants. 21 Chronic bee-paralysis virus-associated satellite virus subgroup: Particles of 12 nm containing three molecules of RNA totaling 1,000 nucleotides. Infects animals. 6. Satellite nucleic acids with positive-sense ssRNA genomes Large linear mRNA satellites: Encode a non-structural protein. Rarely modify disease. Linear RNA of 800 to 1,500 nucleotides (e.g., arabis mosaic virus large satellite RNA). Infects plants. 7. Satellite nucleic acids with negative-sense ssRNA genomes Deltavirus—hepatitis delta virus. Enveloped isometric particle of 36 nm with one circular RNA molecule of 1,700 nucleotides. Dependent on hepatitis B virus, which contributes the envelope, but expresses two delta antigen proteins that form the core. RNA has selfcleaving (ribozyme) activity. May exacerbate hepatitis B virus infection. Infects humans. 8. Satellite nucleic acids with ssRNA genomes (unclassified as make no mRNA) Have no functional ORF. Small linear RNA satellites: Linear RNA of 340 nucleotides (e.g., cucumber mosaic virus satellite RNA). Infects plants. Circular RNA satellites: Covalently closed circular RNA of usually 350 nucleotides. Smallest is 220 nucleotides. All form a double-stranded rodlike viroid RNA. This is encapsidated. Also called virusoids. Have self-cleaving (ribozyme) activity. Five types, all with sobemovirus helpers (e.g., velvet tobacco mottle virus satellite RNA). Infects plants. 22