Particle Image Velocimetry Measurements of Turbulent Flow
advertisement

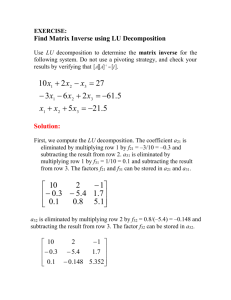
Particle Image Velocimetry Measurements of Turbulent Flow through a Rod Bundle B. L. Smith Experimental Fluid Dynamics Laboratory Utah State University 10 October, 2005 Prepared for U. S. Department of Energy Office of Nuclear Energy, Science and Technology Under DoE Idaho Operations Office Contract DE-AC07-05ID14517 Abstract Distributions of velocity and turbulence have been measured in a rod bundle of two parallel rods downstream of a spacer grid using a twodimensional particle image velocimetry (PIV) system. The measurements were taken in a matched-index-of-refraction (MIR) facility, which allowed the measurement of complete two-dimensional planes surrounding the model fuel rods. The rods were spaced at a pitchto-diameter and wall-to-diameter ratio of P/D = W/D = 1.21. Due to the length of the test section, upstream and down stream measurements were taken at each spanwise location. A FORTRAN code was developed to integrate the two measurement zones into a single velocity field and combine the 2D planes into a 3D velocity field. Introduction The safe, reliable, and efficient operation of nuclear reactors is dependant on the ability to accurately predict velocity and temperature distributions in the flow inside coolant channels. A frequently used fuel element geometry in nuclear reactors is the rod bundle. The development of predictive capabilities for flow through rod bundles requires detailed experimental data on the flow distribution and on the distribution of turbulence intensities. The work presented herein was performed in support of the U.S. / Korean International Nuclear Energy Research Initiative (KNERI) project entitled “Advanced computational thermal fluid physics (CTFP) and its assessment for light water reactors and super-critical reactors.” The goal of the experimental portion of the project is to answer the scientific needs, guide code development and assess code capabilities for treating the generic forced convection problems in Advanced Light Water Reactors (ALWRs) and Super-Critical Reactors (SCRs). The current experiment consists of two parallel rods, representative of a small section of a proposed reactor core. The data provide benchmark velocity and turbulence measurements for the portion of the study dwelling on forced convection in complex reactor geometries. For the representative geometry, the experimental model provides a generic simulation of flow along fuel rods separated by periodic grid spacers, as in an SCR concept. Apparatus and Experimental Technique The experiment was conducted in the matched-index-of-refraction (MIR) flow system at the Idaho National Laboratory (INL). The MIR facility is the largest of its kind in the world, allowing for observation of large-scale models. The positioning of the current model in the MIR system is illustrated in Figure 1. 2 Figure 1. MIR flow system with auxiliary flow loop for model flow control. Nominal Model Design The model represents two fuel rods from a reactor core geometry. It was selected to include flow features of the thermal SCWR concepts suggested by Forschungszentrum Karlsruhe, INL, and Professor Oka, University of Tokyo. The geometry is scaled to be six to seven times larger than typical fuel pins, as depicted in Figure 2. The rod diameter is 2.50 inches (63.5 mm) and the axial pitch of the grid spacers is 17.5 inches (444.5 mm). The outer diameter of the spacers is 3.025 inches (76.8 mm). The inside diameter is 2.85 inches (72.4 mm). The rods are each supported by two half-sphere protuberances and a rod-centering device. Consequently, the cross-section is a rectangular flow channel with nominal dimensions of 3.025 inches by 6.050 inches (76.8 mm by 153.7 mm). The pitch-to-diameter and wall-to-diameter ratio are then P/D = W/D = 1.21 for the simulated fuel rods. The proposed cross-section as described is illustrated in Figure 3. In the axial direction, the model is made up of six geometrically identical sections to produce streamwise-periodic flow conditions, as shown in Figure 4. The model is constructed of a semi-transparent plastic, with the exception of the fourth section, shown in green, which is constructed of fused quartz. At 23.3o C the quartz has the same index-of-refraction as Penreco Drakeol #5 oil, the fluid used in the experiment. The fourth section is referred to as the observation or measurement section where non-intrusive measurements can be taken. 3 Figure 2. SCWR fuel assembly in a square configuration. Figure 3. Nominal model cross-section. 4 Figure 4. Model as built. Fabricated Model The model as fabricated and installed in the MIR flow facility differs slightly from the nominal model design. At the time of this writing there are several unknown dimensions. The system apparently is not exactly symmetrical in the horizontal or vertical directions. At the upstream end of the observation section, the top grid spacer is made of plastic, while the lower one is made of quartz. Since the quartz is stiff and brittle and the plastic is more flexible, the upper spacer may deform and allow the upper rod to settle to a lower elevation. The rods are not perfectly aligned side-to-side nor are they equally spaced vertically in the channel. The variations in the streamwise direction should be measured. The origin of the data set is based on the actual position of the rods and spacers. The streamwise origin (x = 0) is the downstream edge of the upstream spacer. The cross-stream origin (y = 0) is directly between the two rods. The spanwise origin (z = 0) is centered on the lower rod. Measurements Measurements of velocity components are made using a state-of-the-art Particle Image Velocimetry (PIV) system by LaVision Inc. PIV generates a planar two-component velocity vector field at an instant in time. Each measurement requires a pair of digital images separated by a known time increment. The velocity is calculated by correlating the two images. The flow is seeded with ten-micron diameter silver-coated glass spheres. The seeds are chosen to be approximately neutrally buoyant, allowing them to closely follow the flow. The seeds are illuminated by a laser sheet during the capture of each image. Cross-correlations are performed on small subregions (interrogation windows) of the image pairs to determine the most likely velocity vector in the plane for that sub region. The interrogation window initially consists of 64 pixels in each direction and these may overlap one another by fifty percent. The camera is placed perpendicularly to the laser sheet in the fashion shown in Figure 5. 5 Figure 5. General 2-D PIV arrangement. The resolution and accuracy of the result can be improved by shifting the second window in the estimated direction of the velocity vector by a known amount. A first pass without a shift provides the estimate of how much to displace the second window on the second pass. Multiple passes make it possible to reduce the interrogation window to 16 pixels, quadrupling the spatial resolution. The bias uncertainty of each instantaneous PIV measurement is estimated similarly as in [2] by noting that PIV measurements are made by determining the displacement of particles over a set time interval. Specifically, sL0 , u tLI where u is a general velocity component, t is the time interval between the laser pulses, s is the particle displacement from the correlation algorithm, LO is the width of the camera view in the object plane in physical coordinates and LI is the width of the digital image in pixels (the streamwise direction was chosen). Assuming independent samples (as verified below), the bias error of the measured velocity is related to the elementary bias errors based on the sensitivity coefficients: 2 2 u 2 2 u 2 2 u 2 u 2 2 Bu Bs Bt BL0 BL I s t L0 LI The magnitudes and bias uncertainties of each of these quantities are provided in the table below. The velocity bias uncertainty based on these values was computed to be Bu = 0.0183 m/s. The bias in v is similar. Variable Magnitude Bias, Bi Lo (m) 0.25 2E-6 LI [pixels] 1376 0.5 3E-4 1.4E-9 t (s) 10 (typical) 0.03 s (pixel) 6 In addition to the bias error on the instantaneous velocity, the fluctuating, turbulent, velocity field results in a precision error on the mean value. For a 95% confidence interval, the precision error is u Pu 1.96 rms N where N is the number of independent samples (400) and urms is the root-mean-square of the velocity fluctuations, and 1.96 is the multiplier for a 95% confidence interval. The precision uncertainty is thus 0.098urms. Typical values of urms are 0.3 m/s, and in some regions near the m/s. Using the larger value, one finds the precision error spacers, the rms level is as high as 0.5 is about 0.0054 m/s, which is not significant compared to the bias error. Two groups of measurements were made. Most measurements were made with a large field of view using a 50 mm camera lens; these are described below. Subsequently a second set of data was acquired with a 105-mm lens on two very small fields of view (at z = 0) in between the rods, 1) just downstream of a spacer and 2) directly in between two sets of spacers. Very high spatial resolution was achieved, with the entire vertical extent of the camera view spread over 18 mm. An example of one raw-data image from this data set is shown in Figure 6. The red spots are seed particles with reflections bright enough to saturate the camera sensor. When acquiring these data, the seeding density used was the same as the wide-angle shots described below. As a result, the number of pixels from one seed image to the next is much larger than the wide-angle data. It is therefore necessary to use very large interrogation regions to process the data reliably, and doing so would render the vector resolution of this measurement undesirable. Processing with a final interrogation window of 16 pixels generates a large number of invalid vectors, most of which the PIV software detects and removes. However, a small number of invalid vectors can have a very large impact on rms calculations. Therefore, we have decided that the rms results should not be used. The time-mean flow field of the flow downstream of the spacer is shown in Figure 7. The wake of the spacers is clearly visible just downstream from where the two spacers meet. 7 Figure 6. Raw data image from a high resolution case just downstream of a spacer with flow from right to left. The upper spacer (plastic) is clearly visible. The lower spacer, which is quartz and is shifted slightly upstream, is faintly visible. Figure 7. Time-averaged vector field of the flow in the vicinity of the rod spacers. Flow inside the upper spacer cannot be measured since the plastic spacer obscures the camera view (i.e. refractive index of plastic does not match the oil). Flow inside the lower quartz spacer has been measured successfully. For the wide-angle set of data (50-mm lens), due to the length of the observation section, the PIV camera was not able to view the entire section at once. Therefore, flow through the section was captured by taking data at an upstream station, moving the camera downstream and taking additional data at the downstream station. Combining the data from the two stations then created a complete picture of the observation section. The data in the region where the upstream and downstream images overlapped were averaged. Data were acquired in this fashion for 33 spanwise (z) planes. At each position (x,y,z) 400 samples of the instantaneous u and v velocities were collected, from which the flow statistics were calculated. The 33 resulting planes were then combined into one file, which documented the flow through the entire 3D test section. The manner in which the upstream and downstream data were combined is described in Appendix A. For each plane, 400 instantaneous flow fields were acquired at a rate of four per second. This rate was based on a hand calculation of the lowest frequencies likely to be present in the flow and was validated by acquiring sequences of 400 samples at 1 and 2 Hz and verifying that the values did not change. The FORTRAN code used to average all data and assemble the various planes is provided in Appendix C. 8 Results From the 400 instantaneous u and v velocities collected at each spatial point, the mean streamwise and vertical velocity components were calculated, as well as the in plane Reynolds stresses (normal and shear) and the turbulent kinetic energy. The final data set is a 1.6 million data point, 155 MB ASCII (TechPlot format) file. The file has columns of x, y, z, U, V, uu, v v , uv ,TKE. The streamwise origin (x) is located at the downstream edge of the upstream grid spacer. The domain of the data set extends slightly upstream of the origin. The upstream edge of the downstream grid spacer is then located at x = 400 mm. Statistics The mean axial and vertical velocities were calculated in the FORTRAN code (Appendix C) according to Equations (1) and (2) respectively. The number N is the number of valid vectors returned by the PIV system. In the PIV software there are several criteria used to determine the validity of a calculated vector. U 1 N ui N i1 (1) V 1 N vi N i1 (2) In the case of an invalid vector the value of the vector is returned as exactly zero. Also in the software, masks were applied to the portions of the image where the solid model was present, i.e., rods, spacers, walls. The velocity was set identically to zero in these areas. As the instantaneous velocities at each point were summed the number of non-zero values was counted, giving the value of N at that point. If the number of valid vectors at a point was found to less than half the total number of vectors the mean velocity at that point was set to zero. With the mean velocities known, the instantaneous velocities were calculated according to Eqs. (3) and (4), from which the Reynolds stresses were calculated according to Eqs. (5), (6) and (7). With the Reynolds stresses known at each point, the "turbulence kinetic energy" was calculated according to Eq. (8). u i ui U, i 1,N (3) v i v i V, i 1,N (4) u u v v u v 1 N 1 N 1 N N ui ui (5) i 1 N vi vi (6) i 1 N ui vi i 1 9 (7) TKE 1 u u v v 2 (8) Flow Results The bulk velocity at each streamwise plane was calculated by averaging all non-zero velocities in that plane. The resulting axial variation is shown by the lower line in Figure 11. However, there is considerable uncertainty in the velocities near the walls due to the finite thickness of the laser sheet in the PIV system. In the current model, the back wall, i.e. z = 40 mm, is not made of fused quartz. Additionally, O-rings used to assemble the model interfere with the laser as the walls are approached. Despite the well matched indices of refraction for the model material and the fluid, the data at the back of the model (for which the camera looks through the rods) is more scattered. Consequently, we calculate the bulk velocity by averaging only the axial velocities where z ≥ 0, the region nearest the camera. The resulting bulk velocity calculation is shown by the upper line in Figure 11. Clearly this value should not vary with x, and all variations therefore represent uncertainty in the measurement. Based on the two flow meters installed in the MIR in the loops providing flow to the model, the bulk velocity is 4.46 m/s, which is 5% larger than our measurement. Figure 8. Streamwise (U) velocity on several evenly-spaced y-z planes. 10 PIV data taken for the KNERI project in the MIR lab at INL, Idaho Falls, Idaho Mean V velocity May, 2005 V: -0.5 -0.4 -0.3 -0.2 -0.1 0.0 0.1 0.2 0.3 0.4 0.5 321 streamwise planes Range: X = -16.7mm to 410.4mm Origin X: Spacer edge Y & Z: Symmetry Planes 100 0 y 50 -50 Velocity in meters per second Dimensions in millimeters -100 x -40 0 40 z Figure 9. Cross-stream (V) velocity on several evenly-spaced y-z planes. Figure 10. "Turbulence kinetic energy" velocity on several evenly-spaced y-z planes. 11 The mean axial velocity shown in Figure 12 is normalized by the average bulk velocity for the entire volume. Due to the smaller cross section flow path in the area of the grid spacers the mean axial velocity U is highest just as the flow leaves the spacer, as seen in the right most plane of Figure 8. The mean vertical velocity and "turbulent kinetic energy," shown in Figures 9 and 10 respectively, are very small throughout the flow except in the region very near the spacers. As the cross-sectional area of the flow suddenly increases, the maximum axial velocity decreases downstream of the grid spacer. The normalized axial velocity (U/Ubulk) shows higher relative velocities everywhere compared to other studies in similar fuel rod configurations. For the configuration of Rehme, the largest normalized velocity reported is 1.3 [1]. However, the ratio of cross-sectional area in the spacer to the cross-sectional area out of the spacer is smaller for the present configuration than for the configuration in [1]. The greater reduction in crosssectional area would cause the flow to reach higher velocities. The geometry of the spacer is such that flow turbulence would also be increased over the smoother geometry of [1]. As shown in Figure 13, the maximum axial mean velocity decreases rapidly in the first 50 mm after the spacer, then decreases at a nearly constant rate. As the next periodic spacer is approached, the maximum velocity levels off and begins to increase as it approaches the smaller cross-sectional area. Three cross-stream planes (y,z) of data at reduced resolution (for the sake of space) are provided in the Appendix B. These same planes are posted in text files on the EFDL website [3] at full resolution. Conclusions Velocity and turbulence distributions have been obtained for a rod bundle of two parallel rods separated by grid spacers using a PIV system. The resulting data were analyzed to produce mean axial and vertical velocity fields as well as turbulence data. The final results including mass flow rate and bulk velocity are in reasonable agreement with the values measured using the MIR lab equipment. The data provide an opportunity to test turbulence models for the given geometry. 12 Figure 11. Streamwise bulk velocity variation. 13 Figure 12. Streamwise velocity normalized by the bulk velocity. 14 Figure 13. Maximum normalized streamwise velocity (Umax/Ubulk) as a function of downstream distance. References [1] [2] [3] Rehme, K., Experimental Investigation of the Redistribution of Turbulent Flow in a Rod Bundle Downstream of a Spacer Grid, Proceedings of the Fifth International Topical Meeting on Reactor Thermal Hydaulics NURETH-5, 1992. Adeyinka, O. B., and Naterer, G. F., Experimental Uncertainty of Measured Entropy Production with Pulsed Laser PIV and Planar Laser Induced Fluorescence, Int. J. Heat and Mass 48 pp. 1450-1461, 2005. Smith, B. L., Experimental Fluid Dynamics Laboratory website, 2005. http://www.mae.usu.edu/faculty/bsmith/EFDL/EFDL.htm. 15 Appendix A Overlap Region The overlap region was determined by correlating the model geometry with the locations returned by the PIV system. The actual geometry of the model is such that the axial distance between the spacers is approximately 400 mm. The coordinates assigned to the upstream and downstream images by the PIV system are shown in Figures 14 and 15, respectively. To correlate the two coordinate systems a new origin was chosen. The x-origin for the presented data was chosen to be the downstream side of the upper upstream spacer, as indicated by x = 0 in Figure 16. Since the PIV system defaults to flow moving from left to right, the first change was to invert the sign on all x-coordinates. Then, for every point in the upstream image, 28.9 mm was added to the x-coordinate to align it with the new origin. For every point in the downstream image, 190 mm was added to the x-coordinate to align the downstream spacer to its proper location, 400 mm from the upstream spacer. Figure 14. Coordinates for the upstream image. Figure 15. Coordinates for the downstream image. 16 Figure 16. Correlation of the coordinate systems. From the values in Figure Error! Reference source not found. the span of the overlap region was found to be 104.9 mm. Starting 144.39 mm downstream from the upstream spacer, it extended to 249.3 mm. In the non-overlap regions the values of the variables calculated from the PIV results were used. In the overlap region the upstream and downstream values were averaged. 17 Appendix B Selected y-z data planes. The columns are x [mm], y[mm], z[mm], U[m/s], V[m/s], uu[m2/s2], v v [m2/s2], uv [m /s ],TKE [m2/s2]. The streamwise origin (x) is located at the downstream edge of the upstream grid spacer. The domain of the data set extends slightly upstream of the origin. The upstream edge of the downstream grid spacer is then located at x = 400 mm. 2 2 X=32mm (Reduced Data Set) y 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 gf-72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 z 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 U 0.0000 6.2933 5.9315 5.0417 4.7003 2.9336 0.0000 0.0000 0.0000 0.0000 3.0022 4.5596 5.5302 6.6964 7.1146 7.1544 6.6015 4.8936 3.3168 2.5622 0.0000 0.0000 0.0000 0.0000 0.0000 4.9647 5.1210 6.5332 6.2036 0.0000 0.0000 5.0944 4.9067 0.3890 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 4.7220 6.6014 5.9471 5.0018 1.5964 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 4.7377 5.1086 0.0000 0.0000 4.4561 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 V 0.0000 -0.0393 -0.0247 0.0961 0.0709 -0.1310 0.0000 0.0000 0.0000 0.0000 0.4692 0.4773 0.2382 0.1724 0.1118 0.0562 -0.0024 0.1095 0.0168 0.0928 0.0000 0.0000 0.0000 0.0000 0.0000 0.1813 0.1023 0.1245 0.0584 0.0000 0.0000 -0.0592 0.2063 0.0459 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0841 0.1742 0.0784 0.0682 0.0762 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0980 0.0937 0.0000 0.0000 0.0443 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 18 u'u' v'v' 0.0000 0.1466 0.5212 0.3863 0.5920 3.0700 0.0000 0.0000 0.0000 0.0000 1.4251 0.4698 0.5146 0.2112 0.1455 0.1415 0.3152 0.6833 0.5224 0.9894 0.0000 0.0000 0.0000 0.0000 0.0000 0.3475 0.4735 0.1879 0.1771 0.0000 0.0000 0.3759 0.6838 1.3774 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.5107 0.3950 0.7264 0.4061 3.6679 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3909 0.4353 0.0000 0.0000 0.7668 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0773 0.3025 0.3989 0.2930 0.3463 0.0000 0.0000 0.0000 0.0000 0.4243 0.2530 0.2553 0.1369 0.0818 0.0977 0.1846 0.4348 0.3612 0.2692 0.0000 0.0000 0.0000 0.0000 0.0000 0.2505 0.3928 0.1349 0.0722 0.0000 0.0000 0.2771 0.2966 0.0726 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2938 0.1967 0.3809 0.3699 0.1131 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3120 0.2018 0.0000 0.0000 0.2783 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 u'v' TKE 0.0000 0.0104 -0.0379 0.0155 -0.0408 -0.0189 0.0000 0.0000 0.0000 0.0000 0.2165 0.0255 0.0480 0.0469 0.0106 -0.0053 -0.0522 -0.1600 -0.1503 0.0236 0.0000 0.0000 0.0000 0.0000 0.0000 0.0089 0.0314 0.0224 -0.0352 0.0000 0.0000 0.0116 -0.0245 0.0681 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0293 0.0535 -0.0579 0.0216 -0.0704 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0017 -0.0009 0.0000 0.0000 0.0594 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1120 0.4119 0.3926 0.4425 1.7081 0.0000 0.0000 0.0000 0.0000 0.9247 0.3614 0.3849 0.1740 0.1137 0.1196 0.2499 0.5590 0.4418 0.6293 0.0000 0.0000 0.0000 0.0000 0.0000 0.2990 0.4332 0.1614 0.1247 0.0000 0.0000 0.3265 0.4902 0.7250 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.4023 0.2959 0.5536 0.3880 1.8905 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3514 0.3186 0.0000 0.0000 0.5225 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 0.0000 0.0000 0.0000 0.0000 0.0000 3.3484 4.3232 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 3.4123 0.0000 0.0000 3.6635 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 2.3639 3.6795 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 2.0617 0.0000 0.0000 4.5371 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 3.5739 4.5265 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 3.1827 0.0000 0.0000 5.4012 5.0220 4.5945 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 4.9815 0.0000 0.0000 0.0000 0.0000 0.0000 0.0318 0.0956 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0917 0.0000 0.0000 -0.0089 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0136 -0.0338 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0067 0.0000 0.0000 0.0625 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0210 0.0188 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0082 0.0000 0.0000 -0.0182 0.0733 0.0571 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1024 19 0.0000 0.0000 0.0000 0.0000 0.0000 0.4716 0.6893 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.4891 0.0000 0.0000 0.2267 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1020 0.1554 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1277 0.0000 0.0000 0.1599 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2837 0.1851 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1932 0.0000 0.0000 0.1594 0.1246 0.2661 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2357 0.0000 0.0000 0.0000 0.0000 0.0000 0.2668 0.3521 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2671 0.0000 0.0000 0.0901 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0430 0.0854 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0259 0.0000 0.0000 0.0613 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1219 0.1078 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0564 0.0000 0.0000 0.0832 0.1078 0.0802 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0973 0.0000 0.0000 0.0000 0.0000 0.0000 0.0397 0.0577 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0615 0.0000 0.0000 -0.0013 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0179 0.0177 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0020 0.0000 0.0000 0.0176 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0304 0.0321 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0105 0.0000 0.0000 0.0058 0.0145 -0.0017 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0303 0.0000 0.0000 0.0000 0.0000 0.0000 0.3692 0.5207 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3781 0.0000 0.0000 0.1584 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0725 0.1204 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0768 0.0000 0.0000 0.1106 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2028 0.1464 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1248 0.0000 0.0000 0.1213 0.1162 0.1731 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1665 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 6.5702 6.3884 5.1343 4.2235 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 3.9811 4.9830 0.0000 0.0000 6.1615 6.2633 5.4524 4.8078 4.1436 0.0000 0.0000 0.0000 0.0000 2.5043 4.0330 5.6727 6.6458 6.9208 6.9502 6.6530 5.2333 3.3401 2.6816 0.0000 0.0000 0.0000 0.0000 0.0000 4.7700 5.2394 6.4236 6.2781 0.0000 0.0825 0.0159 0.0604 -0.0744 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1242 0.0515 0.0000 0.0000 -0.0275 -0.0590 -0.0098 0.1391 0.0303 0.0000 0.0000 0.0000 0.0000 0.1907 0.2040 0.0970 0.0653 0.0574 0.0342 0.0272 0.0372 0.0721 0.1048 0.0000 0.0000 0.0000 0.0000 0.0000 0.1502 0.0654 0.0651 0.0240 0.0000 20 0.1627 0.2280 0.1769 0.2948 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3827 0.1970 0.0000 0.0000 0.0368 0.0665 0.1452 0.1348 0.2259 0.0000 0.0000 0.0000 0.0000 0.1985 0.2165 0.1877 0.1078 0.0909 0.0813 0.1152 0.3184 0.2678 0.2020 0.0000 0.0000 0.0000 0.0000 0.0000 0.1319 0.1602 0.0734 0.0704 0.0000 0.0486 0.0783 0.1138 0.0770 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0830 0.0886 0.0000 0.0000 0.0191 0.0460 0.1227 0.0786 0.1055 0.0000 0.0000 0.0000 0.0000 0.1384 0.1176 0.0797 0.0509 0.0471 0.0451 0.0610 0.1333 0.1764 0.0981 0.0000 0.0000 0.0000 0.0000 0.0000 0.0718 0.1133 0.0473 0.0211 0.0000 0.0137 -0.0246 -0.0077 -0.0158 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0019 -0.0262 0.0000 0.0000 0.0070 -0.0096 -0.0139 -0.0044 -0.0210 0.0000 0.0000 0.0000 0.0000 0.0220 0.0628 0.0436 0.0226 0.0043 -0.0056 -0.0247 -0.0863 -0.0738 0.0256 0.0000 0.0000 0.0000 0.0000 0.0000 0.0036 0.0214 0.0141 -0.0145 0.0000 0.1056 0.1531 0.1454 0.1859 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2329 0.1428 0.0000 0.0000 0.0280 0.0563 0.1339 0.1067 0.1657 0.0000 0.0000 0.0000 0.0000 0.1685 0.1671 0.1337 0.0794 0.0690 0.0632 0.0881 0.2258 0.2221 0.1501 0.0000 0.0000 0.0000 0.0000 0.0000 0.1018 0.1368 0.0604 0.0458 0.0000 X=80mm (Reduced Data Set) y 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 z 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 U 0.0000 6.0914 5.9251 5.2318 4.6632 3.7005 0.0000 0.0000 0.0000 0.0000 3.5103 4.7015 5.7831 6.6105 6.9882 6.9422 6.2267 4.9221 3.6812 2.8852 0.0000 0.0000 0.0000 0.0000 0.0000 4.9768 5.4867 6.2216 5.8576 0.0000 0.0000 5.0806 4.9567 0.1736 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 5.3038 6.4647 5.8855 5.1696 0.6759 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 4.7472 4.6464 0.0000 0.0000 4.3460 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 4.1095 4.2609 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 3.4008 V 0.0000 0.0048 -0.0173 0.0404 0.0634 0.0111 0.0000 0.0000 0.0000 0.0000 0.1497 0.2425 0.1954 0.1401 0.1248 0.0529 0.0403 0.0619 0.0757 0.1432 0.0000 0.0000 0.0000 0.0000 0.0000 0.0948 0.0531 0.0604 0.0297 0.0000 0.0000 -0.0086 0.0801 -0.0689 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0849 0.1280 0.1266 0.0384 0.1067 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0736 0.0549 0.0000 0.0000 0.0783 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0162 0.0575 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0349 21 u'u' v'v' 0.0000 0.1706 0.2541 0.1793 0.2065 0.7974 0.0000 0.0000 0.0000 0.0000 0.4790 0.4139 0.3294 0.2300 0.1732 0.1665 0.3111 0.3774 0.2700 0.5498 0.0000 0.0000 0.0000 0.0000 0.0000 0.1091 0.1927 0.1631 0.2393 0.0000 0.0000 0.1785 0.2059 0.5024 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.4605 0.3435 0.3298 0.1734 2.3772 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1515 0.3238 0.0000 0.0000 0.1719 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2915 0.1868 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2005 0.0000 0.1174 0.1478 0.1108 0.1010 0.1405 0.0000 0.0000 0.0000 0.0000 0.2238 0.1931 0.1581 0.1163 0.0966 0.1154 0.1966 0.2316 0.1774 0.1161 0.0000 0.0000 0.0000 0.0000 0.0000 0.0814 0.1207 0.1000 0.0957 0.0000 0.0000 0.0960 0.0937 0.0364 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1713 0.1659 0.1829 0.1080 0.0361 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1048 0.1232 0.0000 0.0000 0.0945 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1260 0.1079 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0672 u'v' TKE 0.0000 0.0248 -0.0476 -0.0197 -0.0180 -0.0271 0.0000 0.0000 0.0000 0.0000 0.0950 0.0904 0.0583 0.0432 0.0065 -0.0079 -0.0791 -0.0953 -0.0875 -0.0070 0.0000 0.0000 0.0000 0.0000 0.0000 0.0017 0.0370 0.0198 -0.0402 0.0000 0.0000 0.0090 -0.0100 0.0124 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0698 0.0164 -0.0464 -0.0278 0.0360 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0110 -0.0386 0.0000 0.0000 0.0028 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0286 0.0046 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0155 0.0000 0.1440 0.2009 0.1451 0.1538 0.4689 0.0000 0.0000 0.0000 0.0000 0.3514 0.3035 0.2438 0.1731 0.1349 0.1409 0.2539 0.3045 0.2237 0.3330 0.0000 0.0000 0.0000 0.0000 0.0000 0.0952 0.1567 0.1315 0.1675 0.0000 0.0000 0.1372 0.1498 0.2694 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3159 0.2547 0.2563 0.1407 1.2066 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1281 0.2235 0.0000 0.0000 0.1332 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2088 0.1473 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1339 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 10.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -30.00 -30.00 -30.00 -30.00 0.0000 0.0000 3.7042 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 3.0769 3.7220 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 2.4196 0.0000 0.0000 4.4710 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 4.1979 4.6022 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 3.2437 0.0000 0.0000 5.3156 5.1007 4.6410 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 5.3066 6.4778 6.2228 5.3078 4.2465 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 4.2584 4.7941 0.0000 0.0000 6.0519 6.0806 5.4836 0.0000 0.0000 -0.0082 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0039 0.0044 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0066 0.0000 0.0000 0.0767 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0252 0.0192 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0156 0.0000 0.0000 -0.0185 0.0124 0.0087 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0350 0.0635 0.0541 0.0642 0.0008 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0646 0.0299 0.0000 0.0000 0.0056 -0.0141 0.0148 22 0.0000 0.0000 0.0586 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0689 0.0569 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0631 0.0000 0.0000 0.0630 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1768 0.0852 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1110 0.0000 0.0000 0.0759 0.0476 0.1449 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2315 0.1204 0.1512 0.1011 0.2611 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1950 0.1336 0.0000 0.0000 0.0496 0.0516 0.0806 0.0000 0.0000 0.0153 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0243 0.0212 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0075 0.0000 0.0000 0.0226 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0533 0.0398 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0137 0.0000 0.0000 0.0287 0.0323 0.0297 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0572 0.0509 0.0564 0.0506 0.0428 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0274 0.0359 0.0000 0.0000 0.0232 0.0383 0.0436 0.0000 0.0000 -0.0039 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0173 0.0097 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0003 0.0000 0.0000 0.0060 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0185 0.0113 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0065 0.0000 0.0000 0.0023 -0.0021 -0.0093 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0323 0.0118 -0.0278 -0.0223 -0.0314 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0010 -0.0104 0.0000 0.0000 0.0080 -0.0105 -0.0100 0.0000 0.0000 0.0369 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0466 0.0391 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0353 0.0000 0.0000 0.0428 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1151 0.0625 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0624 0.0000 0.0000 0.0523 0.0400 0.0873 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1444 0.0856 0.1038 0.0758 0.1520 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1112 0.0848 0.0000 0.0000 0.0364 0.0450 0.0621 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 4.7805 4.0187 0.0000 0.0000 0.0000 0.0000 3.2648 4.3891 5.6073 6.4677 6.8070 6.7880 6.2954 5.0067 3.7131 3.0355 0.0000 0.0000 0.0000 0.0000 0.0000 4.8334 5.4430 6.1680 5.9915 0.0000 0.0439 0.0003 0.0000 0.0000 0.0000 0.0000 0.0614 0.1140 0.1087 0.0838 0.0346 0.0052 -0.0073 0.0228 0.1089 0.1218 0.0000 0.0000 0.0000 0.0000 0.0000 0.0403 0.0148 0.0354 0.0182 0.0000 23 0.0588 0.0837 0.0000 0.0000 0.0000 0.0000 0.1045 0.1378 0.1489 0.1001 0.1020 0.1108 0.1325 0.2087 0.1257 0.0819 0.0000 0.0000 0.0000 0.0000 0.0000 0.0592 0.0719 0.0623 0.0798 0.0000 0.0346 0.0423 0.0000 0.0000 0.0000 0.0000 0.0610 0.0439 0.0693 0.0559 0.0466 0.0553 0.0786 0.1079 0.0804 0.0442 0.0000 0.0000 0.0000 0.0000 0.0000 0.0301 0.0357 0.0316 0.0226 0.0000 -0.0138 -0.0314 0.0000 0.0000 0.0000 0.0000 0.0311 0.0331 0.0446 0.0175 -0.0013 -0.0025 -0.0374 -0.0852 -0.0504 -0.0044 0.0000 0.0000 0.0000 0.0000 0.0000 0.0033 0.0174 0.0128 -0.0127 0.0000 0.0467 0.0630 0.0000 0.0000 0.0000 0.0000 0.0827 0.0908 0.1091 0.0780 0.0743 0.0831 0.1055 0.1583 0.1030 0.0630 0.0000 0.0000 0.0000 0.0000 0.0000 0.0447 0.0538 0.0469 0.0512 0.0000 X=320mm (Reduced Data Set) y 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 z 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 30.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 20.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 10.00 U 0.0000 5.5150 5.9515 5.5214 4.7527 3.9927 0.0000 0.0000 0.0000 0.0000 3.9307 4.8879 5.7075 6.0124 6.0603 6.0086 5.7124 5.1398 4.3646 3.6105 0.0000 0.0000 0.0000 0.0000 4.2446 5.0320 5.5925 5.6763 5.0136 0.0000 0.0000 5.2245 4.9404 0.2319 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 5.7205 6.2164 5.8540 5.1953 0.7025 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 4.9841 4.6994 0.0000 0.0000 4.4943 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 5.1719 4.9371 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 V 0.0000 0.0039 -0.0004 -0.0276 0.0417 0.0914 0.0000 0.0000 0.0000 0.0000 -0.0061 -0.0037 0.0897 0.0372 0.0060 -0.0369 -0.0291 -0.0429 0.0317 0.0695 0.0000 0.0000 0.0000 0.0000 0.0142 0.0443 0.0070 0.0108 0.0205 0.0000 0.0000 0.0142 0.0517 0.0322 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0038 0.0344 0.0406 0.0962 0.6166 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0109 -0.0315 0.0000 0.0000 -0.0006 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0494 0.0190 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 24 u'u' v'v' 0.0000 0.1633 0.3785 0.2238 0.3438 0.6707 0.0000 0.0000 0.0000 0.0000 0.6259 0.4074 0.2560 0.2591 0.3141 0.3159 0.2124 0.2826 0.2646 0.4314 0.0000 0.0000 0.0000 0.0000 0.4500 0.2093 0.1331 0.1462 0.2634 0.0000 0.0000 0.2345 0.6372 0.0970 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.4457 0.2454 0.3103 0.4010 1.3642 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3341 0.2886 0.0000 0.0000 0.1628 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3659 0.3104 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0762 0.1001 0.1727 0.1360 0.2366 0.0000 0.0000 0.0000 0.0000 0.2612 0.2492 0.1662 0.1328 0.1504 0.1788 0.1447 0.1730 0.1423 0.1583 0.0000 0.0000 0.0000 0.0000 0.1343 0.1239 0.0852 0.0878 0.1054 0.0000 0.0000 0.0713 0.1699 0.0129 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1563 0.1385 0.1463 0.1618 0.5181 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1137 0.0749 0.0000 0.0000 0.0510 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1026 0.1221 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 u'v' TKE 0.0000 0.0274 0.0834 -0.0062 -0.0611 -0.1200 0.0000 0.0000 0.0000 0.0000 0.0542 0.0670 0.0502 0.0029 0.0159 0.0061 -0.0406 -0.0809 -0.0849 -0.0527 0.0000 0.0000 0.0000 0.0000 0.0414 0.0311 0.0164 -0.0141 -0.0512 0.0000 0.0000 0.0244 -0.0683 0.0012 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0529 0.0116 -0.0516 -0.0413 0.2174 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0375 -0.0284 0.0000 0.0000 -0.0071 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0182 -0.0327 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1197 0.2393 0.1983 0.2399 0.4537 0.0000 0.0000 0.0000 0.0000 0.4435 0.3283 0.2111 0.1959 0.2323 0.2474 0.1786 0.2278 0.2035 0.2949 0.0000 0.0000 0.0000 0.0000 0.2921 0.1666 0.1091 0.1170 0.1844 0.0000 0.0000 0.1529 0.4036 0.0550 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3010 0.1920 0.2283 0.2814 0.9411 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2239 0.1817 0.0000 0.0000 0.1069 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2342 0.2163 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 77.50 72.15 66.81 10.00 10.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -10.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -20.00 -30.00 -30.00 -30.00 3.8797 0.0000 0.0000 3.8495 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 4.0500 4.1409 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 2.9042 0.0000 0.0000 4.2290 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 4.7358 4.7818 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 3.2139 0.0000 0.0000 5.0816 4.9652 3.2668 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 5.2396 5.9903 5.9041 5.2224 4.1590 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 3.8016 4.5024 0.0000 0.0000 5.4910 5.8660 -0.0126 0.0000 0.0000 0.0143 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0178 -0.0174 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0197 0.0000 0.0000 -0.0066 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0198 0.0151 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0171 0.0000 0.0000 -0.0349 -0.0430 -0.1590 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0552 0.0164 -0.0072 -0.0052 -0.1611 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2619 -0.0333 0.0000 0.0000 -0.0188 -0.0491 25 0.3014 0.0000 0.0000 0.0785 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0959 0.0766 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1316 0.0000 0.0000 0.0722 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1506 0.1529 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1585 0.0000 0.0000 0.0906 0.2415 1.8815 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.6065 0.2326 0.1996 0.3486 0.5672 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.5013 0.1654 0.0000 0.0000 0.1039 0.2973 0.0661 0.0000 0.0000 0.0091 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0160 0.0194 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0081 0.0000 0.0000 0.0134 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0415 0.0413 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0215 0.0000 0.0000 0.0848 0.0774 0.1153 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.1413 0.0765 0.0871 0.1410 0.1859 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0829 0.0306 0.0000 0.0000 0.0247 0.0267 -0.0244 0.0000 0.0000 0.0022 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0053 -0.0042 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0150 -0.0067 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0002 0.0000 0.0000 0.0232 0.0152 -0.0257 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0860 0.0090 -0.0120 0.0540 -0.0118 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0133 -0.0106 0.0000 0.0000 0.0090 0.0081 0.1838 0.0000 0.0000 0.0438 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0559 0.0480 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0699 0.0000 0.0000 0.0428 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0960 0.0971 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0900 0.0000 0.0000 0.0877 0.1595 0.9984 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.3739 0.1545 0.1434 0.2448 0.3765 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.2921 0.0980 0.0000 0.0000 0.0643 0.1620 61.46 56.11 50.77 45.42 40.08 34.73 29.38 24.04 18.69 13.34 8.00 2.65 -2.69 -8.04 -13.39 -18.73 -24.08 -29.42 -34.77 -40.12 -45.46 -50.81 -56.15 -61.50 -66.85 -72.19 -77.54 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 -30.00 5.5917 4.9206 4.1722 0.0000 0.0000 0.0000 0.0000 3.8061 4.7337 5.5207 6.0395 6.2254 6.2087 5.9127 5.2872 4.3651 3.2758 0.0000 0.0000 0.0000 0.0000 2.2965 4.4205 5.3043 5.6923 5.1728 0.0000 -0.0774 -0.0764 -0.0748 0.0000 0.0000 0.0000 0.0000 -0.0047 0.0534 0.0678 0.0251 0.0069 -0.0222 -0.0620 -0.0632 -0.0187 0.0179 0.0000 0.0000 0.0000 0.0000 -0.0174 0.0396 0.0250 0.0059 -0.0058 0.0000 26 0.2213 0.2831 0.3913 0.0000 0.0000 0.0000 0.0000 0.2528 0.1790 0.1765 0.1309 0.1391 0.1250 0.1440 0.1876 0.1971 0.3012 0.0000 0.0000 0.0000 0.0000 1.4098 0.2603 0.0938 0.0464 0.0938 0.0000 0.0467 0.0488 0.0630 0.0000 0.0000 0.0000 0.0000 0.0713 0.0633 0.0736 0.0667 0.0600 0.0702 0.0644 0.0911 0.0710 0.0539 0.0000 0.0000 0.0000 0.0000 0.0433 0.0516 0.0459 0.0294 0.0340 0.0000 0.0043 -0.0089 -0.0640 0.0000 0.0000 0.0000 0.0000 0.0571 0.0480 0.0329 0.0146 0.0091 -0.0063 -0.0299 -0.0617 -0.0568 -0.0420 0.0000 0.0000 0.0000 0.0000 0.0293 0.0103 0.0071 -0.0035 -0.0214 0.0000 0.1340 0.1659 0.2272 0.0000 0.0000 0.0000 0.0000 0.1621 0.1212 0.1250 0.0988 0.0996 0.0976 0.1042 0.1394 0.1341 0.1775 0.0000 0.0000 0.0000 0.0000 0.7265 0.1559 0.0698 0.0379 0.0639 0.0000 Appendix C Data Analysis Code The following is a FORTRAN 90/95 code. The code is commented to explain the purpose of each section and clarify input, output, and calculations. 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! ! "stitched" ! ! ! ! ! ! ! ! ! ! ! This code takes directory names containing files of PIV results that have been written in Techplot format with the following variables:'x','y','U','V'. All files in a given directory are results from the same position in space. The results from each directory are averaged to give the average U and V velocities at each point. Velocity fluctuations at each point in space and time are then calculated, i.e. (u' = u - U). The velocity fluctuations are then squared or multiplied then averaged to give the Reynolds Stresses: u'u'(avg), v'v'(avg) and u'v'(avg). The Turbulent Kinetic Engery (TKE)at each point in space is then calculated by TKE = (u'u'(avg) + v'v'(avg))/2. The x and y locations along with the six quantities U,V,u'u'(avg), v'v'(avg), u'v'(avg), and TKE are then written to a file in Techplot format. ! ! ! ! ! ! Note ! that ! Just processor ! will ! DO i 6/12/05 - code essentially finished and ready for testing This particular code is used to combine the results from several different positions in space into a block of 3D data. Data was recorded at two overlapping streamwise locations and 33 spanwise locations. The two streamwise locations are together while the spanwise locations are simply collected into a single file. In the "stitching" portion of the code a sinlge file is created which spans the region covered by the two single regions. In the non-overlap regions the data is simple copied. In the overlap region values from both regions are combined to give an average value, based on position. The new file, which covers the entire space previous covered by two files, is then joined with similar files at other spanwise locations to create the 3D file Adam Richards May 2005 8/09/05 - Comments above revised - TKE calculation corrected in code on char(I): Returns the character cooresponding to the list is processor dependent: for intel: 48=0,49=1,50=2, etc. write the list to the screen to find out what the particular return: = 1,300; Write(*,*)char(i); ENDDO PROGRAM main IMPLICIT NONE ! Variable declarations and descriptions REAL:: xlast, ylast,Usum,Vsum, UVsum REAL:: zpos,z,x_upper,x_lower,delta_x,overlap,zstep 27 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 INTEGER:: i,j,k,m,n,nn,p,q,ipix,jpix,zp,itemp,jtemp,tempint,numfiles,minvv INTEGER:: di, dj, imax, jmax, nx, loc, ny, a, b, nrow, rennum, rennumread INTEGER:: xstar, xfin, ystar, yfin, pick, lext, lname, ldir, indexxl, indexxr, np, deg INTEGER:: indexyb, indexyt, h, t, o, d, c, th, tth, runnum, count, piccount, ntot INTEGER:: lfilein,OV_pix,x_all,iU,ID,qstart,R_freq CHARACTER(len=20) :: filename,list_of_files CHARACTER(len=10) :: tempchar,tempchar1,tempchar2,tempchar3 CHARACTER(len=10) :: tempchar4,tempchar5,tempchar6 CHARACTER(len=10):: junk, nxchar, nychar, chars, view CHARACTER(len=30):: filedir, deffiledir, deffilename, newfold, string1 CHARACTER(len=8):: outa, outb, outc CHARACTER(len=3):: filenum,n_p,dec_pos,int_pos CHARACTER(len=520):: fileprefix1,fileprefix2 CHARACTER(len=30):: filewrite_up,filewrite_dn,filewrite_all,tempin CHARACTER(len=50):: defext, ext, Tech3D_file CHARACTER(len=120):: fileopen REAL, ALLOCATABLE, DIMENSION(:) :: xtemp,ytemp,utemp,vtemp,xU,yU,xD,yD,x,y REAL, ALLOCATABLE, DIMENSION(:,:) :: UavgU,VavgU,u2avgU,v2avgU,uvavgU,TKE_U REAL, ALLOCATABLE, DIMENSION(:,:) :: UavgD,VavgD,u2avgD,v2avgD,uvavgD,TKE_D REAL, ALLOCATABLE, DIMENSION(:,:) :: Uavg,Vavg,u2avg,v2avg,uvavg,TKE REAL, ALLOCATABLE, DIMENSION(:,:,:) :: uinstU,vinstU,uprmU,vprmU REAL, ALLOCATABLE, DIMENSION(:,:,:) :: uinstD,vinstD,uprmD,vprmD INTEGER, ALLOCATABLE, DIMENSION(:,:) :: vvU,vvD,vv ! Establish Defaults Tech3D_file = 'KNERI_3D_MAY_05.dat' deffiledir='F:\KNERI_AR\' deffilename='zp00p0' defext='_PostProc_nomask_PostProc_withmask' ! Collect the file path information Write(*,*) "The filename, filedirectory, and file extension names & cannot contain spaces." Write(*,*) "To accept default values, enter '1'" Write(*,*) "What is the file directory? (",deffiledir(1:11),")" Read(*,*) filedir;If(filedir=='1') Then; filedir=deffiledir; EndIf Do i=1,90; chars=filedir(i:i); If(chars=='') exit; ENDDO; ldir=i-1 !Write(*,*) "What is the root file name?(",deffilename,")" !Read(*,*) filename;If(filename=='1') Then; filename=deffilename; !EndIf Do i=1,90; chars=filename(i:i); If(chars=='') exit; ENDDO; lname=i-1 !Write(*,*) "What is the file extension?(",defext,")" !Read(*,*) ext;If(ext=='1') Then; ext=defext; !EndIf Do i=1,90; chars=ext(i:i); If(chars=='') exit; ENDDO; lext=i-1 ! Information for the overlap region Write(*,*) WRITE(*,*)'What is the x-location of the Upstream spacer? (28.9)' x_upper = 28.9 !READ(*,*)x_upper Write(*,*) WRITE(*,*)'What is the x-location of the Downstream spacer? (-210)' 28 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 x_lower = -210 !READ(*,*)x_lower Write(*,*) WRITE(*,*)'What is the x-distance between the spacers? delta_x = 400 !READ(*,*)delta_x (400)' ! Set the number of pixels for each picture ! This is camera specific ipix = 200;jpix = 150 ! np represents the number of planes in spanwise direction in the 3D space ! np must be less than 99 or this code won't work Write(*,*) WRITE(*,*)'How many planes are to be analyzed? ' READ(*,*)np !np=4 ! nn represents the number of files at each point in space Write(*,*) WRITE(*,*)'How many files are to be read in for each plane? ' READ(*,*)nn !nn=16 ! Set the minimun number of valid vectors required for a spot Write(*,*) WRITE(*,*)'Minimum number of valuid vectors is 25% of total files.' minvv = int(0.25*nn) WRITE(*,*)'Minimum Vectors = ',minvv ! Initial z position (spanwise direction) ! Positive was towards the Bay doors and Negative was ! to the side of the operator station Write(*,*) WRITE(*,*)'What is the initial Z position? ' READ(*,*)zpos !zpos = 0.0 ! Step size inthe Z direction Write(*,*) WRITE(*,*)'What is the step size in Z position? ' READ(*,*)zstep !zstep = -2.5 ! Set reporting frequency for file readin Write(*,*) WRITE(*,*)'How often should the file being read be reported? ' READ(*,*)R_freq !R_freq = 2 ! Allocate the data storage variables ALLOCATE(xtemp(ipix*jpix),ytemp(ipix*jpix),& utemp(ipix*jpix),vtemp(ipix*jpix)) ALLOCATE(UavgU(ipix,jpix),VavgU(ipix,jpix),u2avgU(ipix,jpix), & v2avgU(ipix,jpix),uvavgU(ipix,jpix),TKE_U(ipix,jpix)) ALLOCATE(xU(ipix),yU(jpix),uinstU(ipix,jpix,nn),vinstU(ipix,jpix,nn) & ,uprmU(ipix,jpix,nn),vprmU(ipix,jpix,nn),vvU(ipix,jpix)) ALLOCATE(UavgD(ipix,jpix),VavgD(ipix,jpix),u2avgD(ipix,jpix), & v2avgD(ipix,jpix),uvavgD(ipix,jpix),TKE_D(ipix,jpix)) ALLOCATE(xD(ipix),yD(jpix),uinstD(ipix,jpix,nn),vinstD(ipix,jpix,nn) & ,uprmD(ipix,jpix,nn),vprmD(ipix,jpix,nn),vvD(ipix,jpix)) ! Open the final 3D file for writing open(unit=35,file=Tech3D_file) write(35,'(A25)')'TITLE = "KNERI_3D_MAY_05"' Write(35,'(A63)')'VARIABLES = "x","y","z","U","V","u2","v2","uv","TKE","Val_Vect"' 29 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 Write(*,*)'Press Enter to analyze the data' READ(*,'(a)')tempin ! Iterate through all the planes Do j=1,np WRITE(*,*)'******************************************' WRITE(*,*)'******************************************' Write(*,'(1x,A24,I2)')'Working on plane number ',j Write(*,'(1x,A20,F5.1)')' Z position = ',zpos WRITE(*,*)'******************************************' WRITE(*,*)'******************************************' ! Is the plane in the Positive or Negative region? ! Positive was towards the Bay doors inthe MIR lab ! and Negative was toward the side of the operator station IF(zpos.LT.0.0)THEN n_p = "n" z = abs(zpos) ELSE n_p = "p" z = zpos ENDIF ! Determine the integer part of the current file name If (z<10) Then t=0 o=int(z) int_pos = char(t+48)//char(o+48) Else t=int(z/10) o=z-t*10 int_pos = char(t+48)//char(o+48) EndIf ! Determine the decimal part of the file name dec_pos = char(int((z-int(z))*10)+48) ! Create one file prefix for the upstream file and ! one for the downstream file fileprefix1=filedir(1:ldir)//"z"//n_p(1:1)//int_pos(1:2)//"p" & //dec_pos(1:1)//"up"//ext(1:lext) fileprefix2=filedir(1:ldir)//"z"//n_p(1:1)//int_pos(1:2)//"p" & //dec_pos(1:1)//"down"//ext(1:lext) ! Create the names for the output files filewrite_up = & "z"//n_p(1:1)//int_pos(1:2)//"p"//dec_pos(1:1)//"up"//".dat" filewrite_dn = & "z"//n_p(1:1)//int_pos(1:2)//"p"//dec_pos(1:1)//"down"//".dat" filewrite_all = & "z"//n_p(1:1)//int_pos(1:2)//"p"//dec_pos(1:1)//"all"//".dat" ! Iterate through all the upstream files collecting ! the instantaneous velocities Do i=1,nn ! setup the specific file name by adding the file number to & B00---.dat If (i<10) Then h=0 t=0 30 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 o=i filenum=char(h+48)//char(t+48)//char(o+48) fileopen = & fileprefix1(1:ldir+8+lext)//"\B00"//filenum//".dat" ElseIf (i>=10 .and. i<100) Then h=0 t=int(i/10) o=i-t*10 filenum=char(h+48)//char(t+48)//char(o+48) fileopen = & fileprefix1(1:ldir+8+lext)//"\B00"//filenum//".dat" ElseIf (i>100) Then h=int(i/100) t=int((i-h*100)/10) o=i-h*100-t*10 filenum=char(h+48)//char(t+48)//char(o+48) fileopen = & fileprefix1(1:ldir+8+lext)//"\B00"//filenum//".dat" EndIf ! Open the upstream file OPEN(unit=100,file=fileopen,status='old') IF(mod(i+1,R_freq).EQ.1)THEN Write(*,'(1x,A13,I4,A4,I4,A15)') & 'Opening file ',i,' of ',nn," & \B00"//filenum//".dat" ENDIF ! Read the header (information not needed) READ(100,*)tempchar1 READ(100,*)tempchar2 READ(100,*)tempchar3 ! ! ! From the first file set up the arrays IF(i.EQ.1)THEN ! Read in all the raw data Do k = 1,ipix*jpix READ(100,*)xtemp(k),ytemp(k),utemp(k),vtemp(k) Write(*,222)k,' of ',ipix*jpix ENDDO CLOSE(100) ! Arrange X and Y data into 1D arrays m=0;n=0;xlast=0.0;ylast=0.0 DO p = 1,ipix IF(xtemp(p).NE.xlast)THEN m = m+1 xU(m) = xtemp(p) xlast = xU(m) ENDIF ENDDO DO q = 1,ipix*jpix IF(ytemp(q).NE.ylast)THEN n = n+1 yU(n) = ytemp(q) ylast = yU(n) ENDIF ENDDO All subsequent files ELSE ! Read in all the raw data 31 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 ! Do k = 1,ipix*jpix READ(100,*)xtemp(k),ytemp(k),utemp(k),vtemp(k) Write(*,222)k,' of & ',ipix*jpix,xtemp(k),ytemp(k),utemp(k),vtemp(k) ENDDO CLOSE(100) ENDIF ! Arrange U and V data into 2D arrays m=0;n=1 DO p = 1,ipix*jpix If(m.EQ.ipix)THEN m = 0;n = n+1 ENDIf m = m+1 !**** Changing the sign on all the instantaneous U values ! This is because the flow was right to left and the PIV system ! always assumes flow left to right to be positive UinstU(m,n,i) = -utemp(p) VinstU(m,n,i) = vtemp(p) ENDDO EndDo WRITE(*,*) WRITE(*,*)'UPSTREAM files read in.............' WRITE(*,*) ! Iterate through all the downstream files collecting ! the instantaneous velocities Do i=1,nn ! setup the specific file name by adding the file & number to B00---.dat If (i<10) Then h=0 t=0 o=i filenum=char(h+48)//char(t+48)//char(o+48) fileopen = & fileprefix2(1:ldir+10+lext)//"\B00"//filenum//".dat" ElseIf (i>=10 .and. i<100) Then h=0 t=int(i/10) o=i-t*10 filenum=char(h+48)//char(t+48)//char(o+48) fileopen = & fileprefix2(1:ldir+10+lext)//"\B00"//filenum//".dat" ElseIf (i>100) Then h=int(i/100) t=int((i-h*100)/10) o=i-h*100-t*10 filenum=char(h+48)//char(t+48)//char(o+48) fileopen = & fileprefix2(1:ldir+10+lext)//"\B00"//filenum//".dat" EndIf ! Open the downstream file OPEN(unit=100,file=fileopen,status='old') IF(mod(i+1,R_freq).EQ.1)THEN Write(*,'(1x,A13,I4,A4,I4,A15)') & 'Opening file ',i,' of ',nn," & \B00"//filenum//".dat" ENDIF 32 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 ! ! Write(*,*)fileopen READ(*,'(a)')tempin ! Read the header (information not needed) READ(100,*)tempchar1 READ(100,*)tempchar2 READ(100,*)tempchar3 ! From the first file set up the arrays IF(i.EQ.1)THEN ! Read in all the raw data Do k = 1,ipix*jpix READ(100,*)xtemp(k),ytemp(k),utemp(k),vtemp(k) ENDDO CLOSE(100) ! Arrange data into arrays: X and Y in 1D arrays and & U and V in 2D arrays m=0;n=0;xlast=0.0;ylast=0.0 DO p = 1,ipix IF(xtemp(p).NE.xlast)THEN m = m+1 xD(m) = xtemp(p) xlast = xD(m) ENDIF ENDDO DO q = 1,ipix*jpix IF(ytemp(q).NE.ylast)THEN n = n+1 yD(n) = ytemp(q) ylast = yD(n) ENDIF ENDDO ELSE ! Read in all the raw data DO k = 1,ipix*jpix READ(100,*)xtemp(k),ytemp(k),utemp(k),vtemp(k) ENDDO CLOSE(100) ENDIF !**** m=0;n=1 DO p = 1,ipix*jpix IF(m.EQ.ipix)THEN m = 0;n = n+1 ENDIf m = m+1 Changing the sign on all the instantaneous U values UinstD(m,n,i) = -utemp(p) VinstD(m,n,i) = vtemp(p) ENDDO ENDDO WRITE(*,*) WRITE(*,*)'DOWNSTREAM files read in.............' ! *** Change the x locations on the files to a relative value ! x = 0 is at the downstream side of the upstream spacer DO p = 1,ipix xU(p) = -xU(p) + x_upper xD(p) = -xD(p) + x_lower + delta_x 33 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464 465 466 467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483 484 485 486 487 488 ENDDO ! Determine the length of the overlap region IF(j.EQ.1)THEN OV_pix = 0 qstart = 1 DO p = 1,ipix DO q = qstart,ipix-1 IF((xU(p).LE.xD(q)).AND.(xU(p).GE.xD(q+1)))THEN OV_pix = OV_pix qstart = q ENDIF ENDDO ENDDO + 1 x_all = ipix*2-OV_pix ! Write(*,*)'Total number of pixels = ',x_all Write(35,'(A7,I3,A3,I3,A3,I3,A8)') & 'ZONE I=',x_all,',J=',jpix,',K=',np,',F=POINT' ENDIF ALLOCATE(x(x_all),y(jpix),vv(x_all,jpix),Uavg(x_all,jpix), & Vavg(x_all,jpix),u2avg(x_all,jpix),v2avg(x_all,jpix), & uvavg(x_all,jpix),TKE(x_all,jpix)) ! Calculate averages at the Upstream location WRITE(*,*) WRITE(*,*)'Performing UPSTREAM calculations..............' uprmU = 0.0; vprmU = 0.0 DO m=1,ipix DO n = 1,jpix ! Calculate Uavg and Vavg at the & Upstream location vvU = 0; Usum = 0.0; Vsum = 0.0 DO k = 1,nn IF((uinstU(m,n,k).NE.0.0) & .OR.(vinstU(m,n,k).NE.0.0))THEN vvU(m,n) = vvU(m,n)+1 Usum = Usum + uinstU(m,n,k) Vsum = Vsum + vinstU(m,n,k) EndIf ENDDO If(vvU(m,n).LT.minvv)THEN UavgU(m,n) = 0.0; VavgU(m,n) = 0.0 ELSE UavgU(m,n) = Usum/vvU(m,n); VavgU(m,n) = & Vsum/vvU(m,n) ENDIF ! Calculate U prime and V prime at the & Upstream location DO k = 1,nn IF((uinstU(m,n,k).NE.0.0) & .OR.(vinstU(m,n,k).NE.0.0))THEN uprmU(m,n,k) = uinstU(m,n,k) - & UavgU(m,n) vprmU(m,n,k) = vinstU(m,n,k) - & VavgU(m,n) EndIf 34 489 490 491 492 493 494 495 496 497 498 499 500 501 502 503 504 505 506 507 508 509 510 511 512 513 514 515 516 517 518 519 520 521 522 523 524 525 526 527 528 529 530 531 532 533 534 535 536 537 538 539 540 541 542 543 544 545 546 547 548 549 550 ENDDO Usum = 0.0; Vsum = 0.0; UVsum = 0.0 DO k = 1,nn IF(vvU(m,n).GE.minvv)THEN Usum = Usum + & uprmU(m,n,k)*uprmU(m,n,k) Vsum = Vsum + & vprmU(m,n,k)*vprmU(m,n,k) UVsum = UVsum + & uprmU(m,n,k)*vprmU(m,n,k) EndIf ENDDO ! Calculate Reynolds stresses at the & Upstream location If(vvU(m,n).LT.minvv)THEN u2avgU(m,n) = 0.0 v2avgU(m,n) = 0.0 uvavgU(m,n) = 0.0 TKE_U(m,n) = 0.0 ELSE u2avgU(m,n) = Usum / vvU(m,n) v2avgU(m,n) = Vsum / vvU(m,n) uvavgU(m,n) = UVsum / vvU(m,n) TKE_U(m,n) = (u2avgU(m,n)+v2avgU(m,n))*0.5 ENDIF ENDDO ENDDO Write(*,*)'Upstream complete' ! Calculate averages at the Downstream location WRITE(*,*) WRITE(*,*)'Performing DOWNSTREAM calculations..............' uprmD = 0.0; vprmD = 0.0 DO m=1,ipix DO n = 1,jpix ! Calculate Uavg and Vavg at the & Downstream location vvD = 0; Usum = 0.0; Vsum = 0.0 DO k = 1,nn IF((uinstD(m,n,k).NE.0.0) & .OR.(vinstD(m,n,k).NE.0.0))THEN vvD(m,n) = vvD(m,n)+1 Usum = Usum + uinstD(m,n,k) Vsum = Vsum + vinstD(m,n,k) EndIf ENDDO If(vvD(m,n).LT.minvv)THEN UavgD(m,n) = 0.0; VavgD(m,n) = 0.0 ELSE UavgD(m,n) = Usum/vvD(m,n); VavgD(m,n) = & Vsum/vvD(m,n) ENDIF ! Calculate U prime and V prime at & the Downstream location DO k = 1,nn IF((uinstD(m,n,k).NE.0.0) & .OR.(vinstD(m,n,k).NE.0.0))THEN 35 551 552 553 554 555 556 557 558 559 560 561 562 563 564 565 566 567 568 569 570 571 572 573 574 575 576 577 578 579 580 581 582 583 584 585 586 587 588 589 590 591 592 593 594 595 596 597 598 599 600 601 602 603 604 605 606 607 608 609 610 611 612 uprmD(m,n,k) = uinstD(m,n,k) UavgD(m,n) vprmD(m,n,k) = vinstD(m,n,k) VavgD(m,n) & & EndIf ENDDO ! Calculate Reynolds stresses at the & Downstream location Usum = 0.0; Vsum = 0.0; UVsum = 0.0 DO k = 1,nn IF(vvD(m,n).GE.minvv)THEN Usum = Usum + & uprmD(m,n,k)*uprmD(m,n,k) Vsum = Vsum + & vprmD(m,n,k)*vprmD(m,n,k) UVsum = UVsum + & uprmD(m,n,k)*vprmD(m,n,k) EndIf ENDDO If(vvD(m,n).LT.minvv)THEN u2avgD(m,n) = 0.0 v2avgD(m,n) = 0.0 uvavgD(m,n) = 0.0 TKE_D(m,n) = 0.0 ELSE u2avgD(m,n) = Usum / vvD(m,n) v2avgD(m,n) = Vsum / vvD(m,n) uvavgD(m,n) = UVsum / vvD(m,n) TKE_D(m,n) = (u2avgD(m,n)+v2avgD(m,n))*0.5 ENDIF ENDDO ENDDO Write(*,*)'Downstream complete' ! Splice the Upstream and Downstream values ! averagingin the overlap area i=1;iU=200;iD=198 DO WHILE(iD.GE.1) IF(iU.GT.OV_pix-2)THEN x(i) = xU(iU); vv(i,:) = vvU(iU,:); Uavg(i,:) = UavgU(iU,:) Vavg(i,:) = VavgU(iU,:); u2avg(i,:) = u2avgU(iU,:) v2avg(i,:) = v2avgU(iU,:); uvavg(i,:) = uvavgU(iU,:) TKE(i,:) = TKE_U(iU,:) i = i+1; iU = iU-1 ElseIF(iU.GE.3)THEN x(i) = (xU(iU)+xD(iD)+xD(iD-1))/3.0 vv(i,:) = (vvU(iU,:)+vvD(iD,:)+vvD(iD-1,:))/3.0 Uavg(i,:) = (UavgU(iU,:)+UavgD(iD,:)+UavgD(iD-1,:))/3.0 Vavg(i,:) = (VavgU(iU,:)+VavgD(iD,:)+VavgD(iD-1,:))/3.0 u2avg(i,:) = (u2avgU(iU,:)+u2avgD(iD,:)+u2avgD(iD-1,:))/3.0 v2avg(i,:) = (v2avgU(iU,:)+v2avgD(iD,:)+v2avgD(iD-1,:))/3.0 uvavg(i,:) = (uvavgU(iU,:)+uvavgD(iD,:)+uvavgD(iD-1,:))/3.0 TKE(i,:) = (TKE_U(iU,:)+TKE_D(iD,:)+TKE_D(iD-1,:))/3.0 i = i+1; iU = iU-1; iD = iD-1 ELSE 36 613 614 615 616 617 618 619 620 621 622 623 624 625 626 627 628 629 630 631 632 633 634 635 636 637 638 639 640 641 642 643 644 645 646 647 648 649 650 651 652 653 654 655 656 x(i) = xD(iD); vv(i,:) = vvD(iD,:); Uavg(i,:) = UavgD(iD,:) Vavg(i,:) = VavgD(iD,:); u2avg(i,:) = u2avgD(iD,:) v2avg(i,:) = v2avgD(iD,:); uvavg(i,:) = uvavgD(iD,:) TKE(i,:) = TKE_D(iD,:) i = i+1; iD = iD-1 ENDIF ENDDO y = yU !Write a file storing the data at the current plane and & add to the 3D file WRITE(*,*) Write(*,*)'Writing to file: ',filewrite_all open(unit=25,file=filewrite_all) Write(25,'(A48)')'VARIABLES = "x","y","U","V","u2","v2","uv","TKE"' Write(25,'(A7,I3,A3,I3,A8)')'ZONE I=',x_all,',J=',jpix,',F=POINT' DO p = 1,jpix DO i = 1,x_all write(25,'(8F12.6)')x(i),y(p),Uavg(i,p),Vavg(i,p), & u2avg(i,p),v2avg(i,p),uvavg(i,p),TKE(i,p) write(35,'(10F12.6)')x(i),y(p),zpos,Uavg(i,p),Vavg(i,p), & u2avg(i,p),v2avg(i,p),uvavg(i,p),TKE(i,p),vv(i,p) ENDDO EndDo close(25) ! move to the next plane and repeat zpos = zpos + zstep ! Deallocate arrays DEALLOCATE(x,y,vv,Uavg,Vavg,u2avg,v2avg,uvavg,TKE) ! Write(*,*)'Press Enter to continue to next plane' ! READ(*,'(a)')tempin EndDo 222 FORMAT(1x,i6,A4,i6,2f10.3,2f7.3) close(35) END PROGRAM main 37