theoretical simulation of raman spectra of dna bases
advertisement

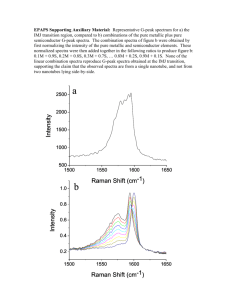
STUDIA UNIVERSITATIS BABES-BOLYAI, PHYSICA, SPECIAL ISSUE, 2003 THEORETICAL SIMULATION OF THE RAMAN SPECTRA OF DNA BASES C. I. Morari, C. M. Muntean National Institute for Research and Development of Isotopic and Molecular Technologies, 400293 ClujNapoca, P.O. 5, Box 700, Romania Abstract Our present study reports the Raman spectra of each separate DNA bases (adenine, cytosine, thymine and guanine). The simulations were performed at ab initio level using the DZ gaussian basis set. The steps of the simulations were the following: (i) optimisation of the geometry for each DNA base; (ii) computing the hessian matrix and the IR spectra for each DNA base; (iii) computing the Raman intensities by using the ab initio polarisabilities. All the calculations were performed using the GAMESS package. Each simulation is reported separately for adenine, cytosine, thymine and guanine. As a first approach for the Raman spectrum of the whole DNA molecule we use the overlap of these four spectra. Introduction DNA is a very important biological molecule since it carries the genetic code. The famous double helix structure has emphasized the relationship between structure and function. In the specific case of DNA, understanding the processes by which specific binding sites are recognized by drugs, mutagens, proteins and other molecules would represent a fundamental step towards understanding its biological activity. Raman spectroscopy was presented as an alternative structural approach for the study of nucleic acids and their complexes, particularly for investigating the aqueous solution structures and the interactions of large oligonucleotides, native nucleic acids and their biological assemblies, like chromosomes and viruses. The high information content of Raman spectra is reflected in the many new insights that have been obtained into the conformation, composition, interaction and functioning of DNA molecules. As a first approach of comparing the experimental results of DNA molecule with theoretical calculations, our present study reports the Raman spectra of each separate DNA base (adenine, cytosine, thymine and guanine) and the overlap of these four spectra. C. I. MORARI, C. M. MUNTEAN Computational methods In order to compute the normal modes of a polyatomic molecule, we have to build the Hessian matrix Fij [1] . The eigenvalues for the vibrational modes are the results of the following system (1) ( F ij ij )l 0 k jk j which can be solved by asking to its determinant to vanish. det(F ij ij k ) 0 (2) The final vibrational frequencies are where k k k 1/ 2 / 2 (3) are the frequencies resulting from the Eq. (1). All the calculation presented below are performed using the GAMESS implementation of this scheme [2]. The structure for all DNA components were optimized using the DZ basis set [3]. The resulting geometries are depicted in Figures 1 and 2. Figure 1: The structures of cytosine (C) and guanine (G).} Figure 2:The structures of adenine (A) and thymine (T) THEORETICAL SIMULATION OF THE RAMAN SPECTRA OF DNA BASES Results The vibrational analysis was performed by computing the Hessian matrix and the force constants for all normal modes of each DNA base. The computed frequencies were scaled with a factor of 0.89 in order to remove the effect of the harmonic oscillator approach upon the computed frequencies [4]. The resulting spectra are given in Figures 3 and 4. The Figure 5 is the overlap for all 4 spectra computed independently. Figure 3: The Raman spectra of cytosine (left) and guanine (right) Figure 4:The Raman spectra of adenine (left) and thymine (right)} Figure 5: Overlap of the spectra given in the Figures 3 and 4. Left: the complete calculation. Right: details for the region 600 cm-1 - 1800 cm-1. C. I. MORARI, C. M. MUNTEAN The significant breathing modes are 681 cm-1 (G) 727 cm-1 (A), 749 cm-1 (T) and 784 cm-1 (C) [5]. These values are reproduced by our calculations (see Figures 3 and 4). The bands at 1304 cm-1 (A, marker "B" in Figure 5), 1376 cm-1 (T,A, G, marker "C" in Figure 5), 1489 cm-1 (G, A, "D" in Figure 5), 1578 cm-1 (G, A, "E" in Figure 5) shows a good correlation with the experimental results. The band located near 1668 cm-1 which was assigned to coupled C=O stretching and N-H deformation modes of T, G and C do exists in the theoretical spectra of thymine (marker "F"). Significant differences between the theoretical and experimental spectra occur for the intensities of the Raman bands (markers "B", "C", and "D", Figure 5.)A possible explanation for these changes are the changes in the polarizability of the whole DNA molecule compared to the polarizabilities of its components. Conclusion An ab initio simulation of the Raman spectra for the DNA bases was performed. The results confirm the previously experimental assignments. Significant difference of the Raman intensity for the simulated and experimental spectra exist. This is a clear indication of the influence of the interactions between base pairs and of the solvent effects upon the computed Raman intensities. References 1. I. N. Le v i ne , "Quantum Chemistry", Prentice Hall, 2000. 2. M.W .S c h mid t, K. K. B ald rid g e, J . A.B o a tz, S.T .Elb er t, M. S. Go rd o n, J .H.J e ns e n, S.Ko se k i, N.Ma t s u na ga , K. A.N g u ye n , S.J . S u, T . L.W i nd u s, M.D up u is , J . A. Mo nt go m er y, J.Comput.Chem. 14, 1347-1363(1993). 3. T .H.Du n ni n g , J r ., P .J . Ha y in "Methods of Electronic Structure Theory", H.F .S h ae fer III, Ed. Plenum Press, N.Y. 1977. 4. A. P . Sco tt, L. Rad o m , J. Phys. Chem. 100, 16502-16513 (1996). 5. C. M. M u nt ea n , "Teza de doctorat", Cluj, 2002.






![supplementary material[R2]](http://s3.studylib.net/store/data/006645029_1-d1fd1feedd0302aa1b544732e09eab5d-300x300.png)