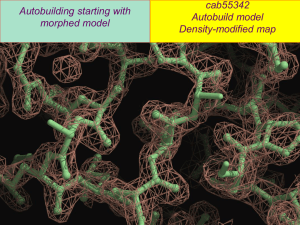
Experimental electron density
advertisement

Crystal Structure of Rac1 in Complex with the Guanine Nucleotide Exchange Region of Tiam1 David K. Worthylake*, Kent L. Rossman†, & John Sondek *†‡ *Department of Pharmacology, †Department of Biochemistry and Biophysics, ‡Lineberger Comprehensive Cancer Center, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599 USA ‡Corresponding author Manuscript W06555A Supplementary Information Supplementary Figure 1 Experimental electron density. The Tiam1 DH domain (yellow), Rac1 (green) and switch regions (red) are superimposed on the experimental electron density (white) generated using density-modified phases and contoured at 1. 1 anomalous differences and model-derived phases were used to generate difference density (red) contoured at 6 and encompassing Met 1224 (at right). This figure was made using SPOCK (http://quorum.tamu.edu/jon/spock/) Supplementary Figure 2 Sequence alignments. a, The all helical secondary structure of the DH domain of Tiam1 is indicated in yellow above its sequence and agrees well with secondary structure elements of other DH domains1-3 highlighted in grey. Tiam1 residues in red italics bury more than 10 Å2 of solvent accessible surface area upon complex formation with Rac1; areas buried are charted in inset. Highly conserved regions among all DH domains are labeled CR1-3 and the majority of Tiam1 residues that interact with Rac1 are localized to CR1 and CR3 and most likely represent a common binding surface for all combinations of G-proteins with Dbl family members. Interacting residues outside the conserved regions localize to sequences within or adjacent to helices 4b, 5, 310-7a, and 9. Residues in the DH domain of Trio that possess significantly broadened amide resonances upon Rac1 addition2 are in black italics and agree well with a subset of the contacts observed in the Tiam1Rac1 complex. Borders of expression constructs are indicated with small arrows, small dots indicate every 10 residues, and lightly shaded italicized residues could not be localized upon structure determination. The secondary structure for Trio was taken from Liu et al. while others were calculated using DSSP4. Twenty-two non-redundant DH domains were aligned using ClustalX5 followed by minimal manual adjustment and only DH domains of known structure are shown. Consensus sequence generated using GCG 6 (plurality, 8; threshold, 1; individual weights, 1). b, Residues in Rac1 that bury more than 10 Å2 upon complex formation with Tiam1 are indicated in bold italics and nomenclature for secondary structure elements derives from the structure of Ras7. Buried residues highlighted red differ between Rac1 and Cdc42 and are therefore likely to be important for dictating specificity between G-proteins and DH domains. Also highlighted are the switch regions that undergo conserved, nucleotide-dependent conformational alteration in G-proteins, as well as, the 21 amino acid insertion unique to Rho family G-proteins, absent in Ras, and involved in some effector interactions. Sequences for 16 Rho family members and Ras were aligned using ClustalX5 and the subset shown. Secondary structure was calculated using DSSP4 1. Aghazadeh, B. et al. Structure and mutagenesis of the Dbl homology domain. Nat Struct Biol 5, 1098-107 (1998). 2. Liu, X. et al. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell 95, 269-77 (1998). 2 3. 4. 5. 6. 7. Soisson, S. M., Nimnual, A. S., Uy, M., Bar-Sagi, D. & Kuriyan, J. Crystal structure of the Dbl and pleckstrin homology domains from the human Son of sevenless protein. Cell 95, 259-68 (1998). Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577-637 (1983). Thompson, J. D., Higgins, D. G. & Gibson, T. J. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties, and weight matrix choice. Nucleic Acids Res. 22, 46734780 (1994). Genetics, Computer & Group. Program Manual for the GCG Package, Version 7, April 1991,575 Science Drive, Madison, Wisconsin, USA 53711. (1991). Pai, E. F. et al. Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene product p21 in the triphosphate conformation. Nature 341, 209-14 (1989). 3