bsc1 - modularity - Protein Evolution (Rob Russell)
advertisement
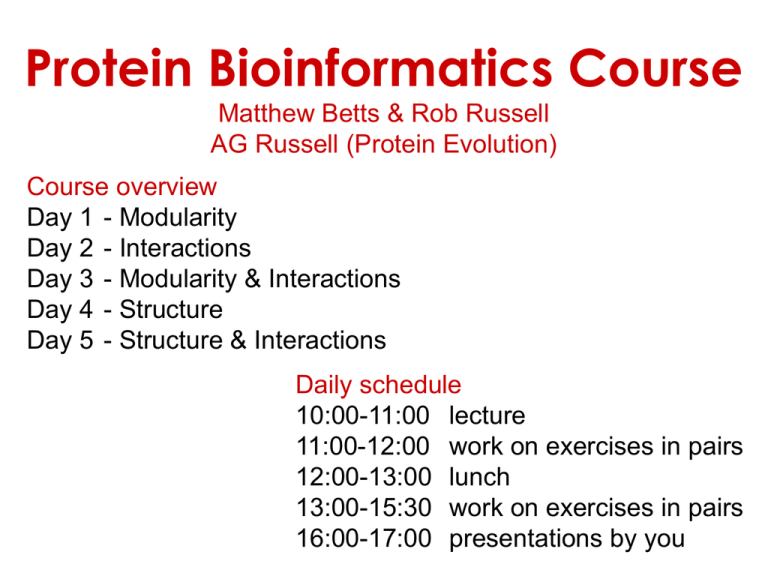
Protein Bioinformatics Course Matthew Betts & Rob Russell AG Russell (Protein Evolution) Course overview Day 1 - Modularity Day 2 - Interactions Day 3 - Modularity & Interactions Day 4 - Structure Day 5 - Structure & Interactions Daily schedule 10:00-11:00 lecture 11:00-12:00 work on exercises in pairs 12:00-13:00 lunch 13:00-15:30 work on exercises in pairs 16:00-17:00 presentations by you Protein Sequence Databases Database Searching • Homologues = proteins with a common ancestor • Homology --> similar function • Sequence similarity --> homology • Find homologues using: • BLAST • Profile Searching www.proteinmodelportal.org Scores and E-values How similar is my sequence to one in the database? • Alignment • Substitution matrix • Gap penalties How much would I expect to get >= this score by chance alone? • cf. random sequences • E = 1: one such match by chance • E < 0.01: significant • Depends on database: • size: larger = better • composition (random assumed) Homology comes in two main types: Orthology and Paralogy What is the difference and why does this matter? Duplication - Speciation Duplication - Paralogues Orthologues - Speciation Paralogues Different Fates Orthologues: • Both copies required (one in each species) • conservation of function (‘same gene’) • adaptation to new environment Easier to transfer knowledge of function between orthologues Paralogues: • Both copies useful • conservation of function • One copy freed from selection • disabled • new function • Different parts of each free from selection • function split between them Assignment of orthology / paralogy can be complicated by: • duplication preceding speciation • lineage-specific deletions of paralogs • complete genome duplications • many-to-one relationship • multi-domain proteins Homology usually found by sequence similarity, but …proteins with dissimilar sequences can still be homologous Betts, Guigo, Agarwal, Russell, EMBO J 2001 Proteins are modular Since the early 1970s it has been observed that protein structures are divided into discrete elements or domains that appear to fold, function and evolve independently. Given a sequence, what should you look for? • Functional domains (Pfam, SMART, COGS, CDD, etc.) • Intrinsic features – – – – Signal peptide, transit peptides (signalP) Transmembrane segments (TMpred, etc) Coiled-coils (coils server) Low complexity regions, disorder (e.g. SEG, disembl) • Hints about structure? Given a sequence, what should you look for? “Low sequence complexity” (Linker regions? Flexible? Junk? Signal peptide (secreted or membrane attached) Transmembrane segment (crosses the membrane) Immunoglobulin domains (bind ligands?) Tyrosine kinase (phosphorylates Tyr) SMART domain ‘bubblegram’ for human fibroblast growth factor (FGF) receptor 1 (type P11362 into web site: smart.embl.de) Protein Modularity • discrete structural and functional units • found in different combinations in different proteins Receptor-related tyrosine-kinase Non-receptor tyrosine-kinases consider separately in predictions Finding Protein Domains • through partial matches to whole sequences: match query sequence: match match • compare to databases of domains (Pfam, SMART, Interpro) • can be separated by: • low-complexity and disordered regions (SEG) • trans-membrane regions (TMAP) • coiled-coils (COILS) Repeat searches using each domain separately 12 000 domain alignments make sequence searching easier WPP domain alignment Alignments provide more information about a protein family and thus allow for more sensitive sequences than a single sequence. Domain alignments also lack low-complexity or disorder (normally) and other domains that can make single sequence searches confusing. Finding domains in a sequence Cryptic domains: at the border of sequence detectability Identified using more sensitive fold recognition methods that use structure to help find weak members of sequence families. If Pfam or SMART or similar do not find a domain, and the region is probably not disordered, then fold recognition might help. Gallego et al, Mol Sys Biol 2010 Domain peptide interactions Recognition of ligands or targeting signals Post-translational modifications Linear motifs Peptides interacting with a common domain often show a common pattern or motif usually 3-8 aas. 3BP1_MOUSE/528-537 APTMPPPLPP PTN8_MOUSE/612-629 IPPPLPERTP “instance” SOS1_HUMAN/1149-1157 VPPPVPPRRR NCF1_HUMAN/359-390 SKPQPAVPPRPSA PEXE_YEAST/85-94 MPPTLPHRDW SH3-interacting motif PxxP “motif” “perpetrator” “victim” Puntervol et al, NAR, 2003; www.elm.org (Eukaryotic Linear Motif DB) Linear motifs versus domains Domains: large globular segments of the proteome that fold into discrete structures and belong in sequence families. Linear motifs: small, non-globular segments that do not adopt a regular structure, and aren’t homologous to each other in the way domains are. Motifs lie in the disordered part of the proteome. Intrinsically unstructured or disordered proteins or protein fragments Disorder predictors (IUPred, RONN, DisORPred, etc) Linear motif mediated interactions are everywhere Include motifs for: • Targeting – e.g. KDEL • Modifications – e.g. phosphorylation • Signaling – e.g. SH3 About 200 are currently known, likely many more still to be discovered Neduva & Russell, Curr. Opin. Biotech, 2006 Finding linear motifs in a sequence Linear motifs are much harder to find than domains. Short (typically < 8AA), simple patterns, e.g. PxxP will occur in most sequences randomly. Long (>30 AA), belong to sequence families that help detect new family members www.russelllab.org/wiki