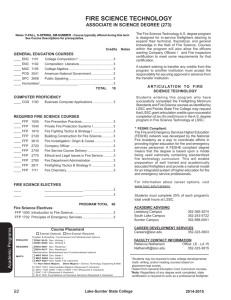
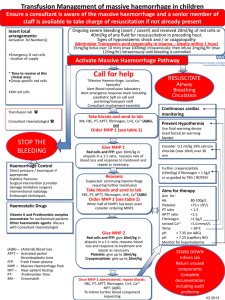
The initial electron density was sufficiently clear to place rigid body
advertisement

Supplementary Experimental Methods Protein expression and purification Recombinant proteins were overexpressed in E. coli strain BL21(DE3) from pET vectors (Novagen) and purified from bacterial lysate by either conventional chromatography (untagged or MBP-tagged ) or by affinity chromatography on Ni-NTA resin (His6-tagged). Wild type Bni1p FH2 domain (residues 1327-1769) and homodimeric point mutants were expressed untagged and His6-tagged, respectively. MBP-tagged tethered FH2 construct was generated by genetically fusing two FH2 domains through an Ala-Met-Gly linker. MBP was removed by thrombin digestion followed by anion-exchange chromatography. Actin was prepared from rabbit muscle acetone powder1, and labeled with pyrene or Tetramethylrhodamine (Molecular Probes) as reported2,3. Structure determination of the FH2-actin complex Diffraction quality The diffraction quality of both our native and selenomethionine-derivatized (SeMet) crystals was limited when using synchrotron radiation in several ways. First, the SeMet crystals were prone to radiation damage. Second, their average diffraction intensity was relatively strong to a Bragg spacing (dmin) of about 4 Å, but suffered a dramatic decrease to the high-resolution limit (2.95 Å—but see below, only data to 3.50 Å were ultimately used in phase determination). Third, the SeMet crystals diffracted isotropically to a dmin of about 4.5 Å, but anisotropically to a dmin of 2.95 Å. Anisotropy was less pronounced but still significant for our highest-quality native dataset for resolution higher than 4 Å. Finally, calculation of Wilson B-values (although limited in its information content, due to the low resolution) indicated a significant degree of disorder (113 Å2 for our native dataset). These features are not atypical for diffraction data of proteins that contain large -helical bundles4-6. Phasing Data were indexed, integrated and scaled using HKL20007. Intensities were converted to structure factor amplitudes and placed on an approximate absolute scale using TRUNCATE from the CCP4 package8,9. We found that a single anomalous dispersion (SAD) experiment was not sufficient to locate selenium sites in our SeMet crystals. However, these sites could be identified successfully using a three-wavelength MAD dataset. Data near the high-resolution limit (2.95 Å) showed limited completeness (30-40 %), due to anisotropy and radiation damage. Because of these shortcomings, only data to 3.5 Å, which coincides roughly with the resolution limit for which anomalous signal was observed, were used for locating and refining selenium sites as well as in density modification. Six selenium sites were identified from the peak and inflection point energy data sets using the Patterson search routine in CNS 1.110. Phases were refined between 35.0 and 3.5 Å using all three SeMet data sets in the program MLPHARE11. The figure-of-merit was 0.36 after phase refinement. Phases were further improved by density modification with histogram matching in the program DM12, resulting in a final overall figure of merit of 0.56. The resulting electron density map was sufficiently clear to place rigid body models of TMR-actin (PDB code 1NWK), including the bound ATP, and the FH2 domain of Bni1p (PDB code 1UX5). To confirm the validity of the SeMet-derived phases, we carried out molecular replacement using TMR-actin (PDB ID code 1nwk, stripped of the Ca2+-ATP) as a search model against the native data set for the actin/FH2 complex in the program AMoRe. The resulting solution coincides with the actin density observed in the SeMet-phased map. In addition, strong density for the missing Ca2+-ATP moiety and the Bni1p FH2 domain were clearly observed in this map. Model refinement The structure was refined in the CNS 1.1 program suite with a random 4.7% subset of all data set aside for Rfree calculation. We corrected the model and refined it against our native dataset using all data between a dmin of 30.0 and 3.05 Å. Iterative cycles of simulated annealing, standard positional and grouped atomic displacement parameter (“B-value”) refinement via a maximum likelihood target were carried out in the program CNS. B-value refinement, although limited in its information content at this resolution, helped to stabilize the overall refinement procedure. The resulting electron density map was reasonable for this type of crystal, but its quality was heterogeneous: some regions had the appearance of a “typical 3 Å structure”, whereas others were significantly worse. Many side chains did not have electron density associated with them. Either pruning these side chains to alanine (for refinement purposes) or setting their occupancy to zero did not lead to improvements in the refinement behavior nor in the resulting electron density. The best agreement between model and data was obtained when all side chains were present, probably because the resulting molecular mask was more accurate and thus the bulk solvent correction more powerful. The B-values for individual subdomains of actin and FH2 in our model range from 91 Å2 to 139 Å2 with an average of 103 Å2, which is typical for this type of crystal (for a recent example of a similar refinement behavior, but at even higher resolution, see Bowman, G.D., O’Donnell, M., & Kuriyan, J., Nature, 2004, 429, 724). TLS-refinement using the program RefMac (G. N. Murshudov, Acta Crystallogr D Biol Crystallogr 53, 240 (1997)) was used to extract TLS parameters for actin and FH2, but did not converge when the model was subdivided into smaller portions. TLS-refinement did not improve the quality of the electron density map, and even led to an increase in the R-factors. A detailed list of average B-values (without TLS-refinement) for the individual actin and FH2 subdomains is presented in the Methods section of the main text to indicate their relative degrees of order. The final model was validated using a composite simulated-annealing omit map calculated in CNS. The omit map was virtually identical wi -weighted 2Fo-Fc electron density map obtained with a maximum-likelihood refinement target and showed contiguous density for all but three isolated residues in our model, residues 230 and 252 in actin, and residue 1488 in the FH2 domain. NMR Analyses of the mDia1 FH2 domain For NMR, the FH2 domain of mDia1 (745-1209) was expressed in 99% D2O M9 media supplemented with 40 mg/l 2-keto-3-methyl -13C, 3,4,4,4-D4-butyric acid initially and 68 mg/l 2-keto-3,3-D2-butyric -4-13C- acid at induction to achieve U-[15N, 2H] Ile 1-[13CH3] and Leu, Val-[13CH3/12CD3] labeling13. NMR experiments were carried out at 25 °C on a Varian Inova 800 MHz spectrometer. Samples were in D2O buffer containing 10 mM Hepes pH 7.0, 150 mM KCl, 0.1 mM CaCl2, 1 mM DTT. 1 H-13C TROSY HMQC data were acquired on free U-[15N,2H], Ile 1-[13CH3], Leu, Val -[13CH3/12CD3]-labeled mDia1 FH2 domain (100 M), and in the presence of 25 M TMR-actin. Data processing and analysis was carried out using NMRPipe and NMRview, respectively14,15. References 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. 14. 15. Spudich, J. A. & Watt, S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem 246, 4866-71. (1971). Pollard, T. D. & Cooper, J. A. Quantitative analysis of the effect of Acanthamoeba profilin on actin filament nucleation and elongation. Biochemistry 23, 6631-41. (1984). Otterbein, L. R., Graceffa, P. & Dominguez, R. The crystal structure of uncomplexed actin in the ADP state. Science 293, 708-11 (2001). Xu, Y. et al. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell 116, 711-23 (2004). Chen, X. et al. Three-dimensional structure of the complexin/SNARE complex. Neuron 33, 397-409 (2002). Michaely, P., Tomchick, D. R., Machius, M. & Anderson, R. G. Crystal structure of a 12 ANK repeat stack from human ankyrinR. EMBO J. 21, 6387-96 (2002). Otwinowski, Z. & Minor, W. Processing X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307-326 (1997). French, G. S. & Wilson, K. S. On the treatment of negative intensity observations. Acta Crystallogr. A 34, 517-525 (1978). COLLABORATIVE COMPUTATIONAL PROJECT, N. The CCP4 Suite: Programs for Protein Crystallography. Acta Cryst. D50, 760-763 (1994). Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905-21 (1998). Otwinowski, Z. in Isomorphous Replacement and Anomalous Scattering (ed. Leslie, A. G. W.) 80-86 (Science & Engineering Research Council, 1991). Cowtan, K. & Main, P. Miscellaneous algorithms for density modification. Acta Crystallogr. D54, 487-493 (1998). Tugarinov, V. & Kay, L. E. Ile, Leu, and Val methyl assignments of the 723residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc 125, 13868-78 (2003). Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277-293 (1995). Johnson, B. A. & Blevins, R. A. NMRView: a computer program for the visualization and analysis of NMR data. J. Biomol. NMR 4, 603-614 (1994).