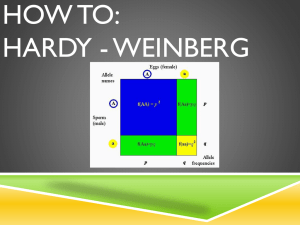
Answer Key - Population Genetics Questions

Practice Problems: Population Genetics
1.
The gamma globulin of human blood serum exists in two forms, Gm(a+) and
Gm(a-), spedified respectively by an autosomal dominant gene Gm(a+) and its recessive allele Gm(a-). Broman et al. (1963) recorded the tabulated phenotypic frequencies in three Swedish populations. Assuming the populations were at Hardy-Weinberg equilibrium, calculate the frequency of heterozygotes in each population.
Region No. Tested Phenotype %
Norbotten County
Stockholm city and rural district
139
509
Gm(a+)
55.40
57.76
Gm(a-)
44.60
42.24
Malmohus and 293 54.95 45.05
Kristianstad counties
Since the populations ARE in equilibrium, this problem can be solved by defining the proportion of individuals displaying the recessive phenotype (GM (A-) as q 2 . You can then calculate q , and using 1q = p , you can calculate p . Finally, the frequency of heterozygotes in a population in equilibrium is 2 pq .
Norbotten County: q2 = 0.446 q = 0.668 p = 0.332
2 pq = 0.444
Stockholm
Malmohus
2
2 pq pq
= 0.455
= 0.442
2.
Among 2,820 Shorthorn cattle, 260 are white, 1,430 are red, and 1,130 are roan. Is this consistent with the assumption that the traits are controlled by a single pair of autosomal alleles and that mating has been at random for this allele pair?
To determine if a population is in equilibrium, you must calculate the allele frequencies, use those allele frequencies to calculate the expected number of white, red and roan cattle, and then use chi-square analysis to compare the expected numbers with the observed numbers.
First: R 1 = red allele
R 2 = white allele
Allele frequencies: p = f(R1) = (1430 + ½(1130)) / 2820 = 0.71 q = 0.29
Expected # Red = p 2 x total cattle =( 0.71
2 )( 2820) = 1421.562
Expected # Roan = 2 pq x total cattle = 2(0.71)(0.29)(2820) = 1161.276
Expected # White = q 2 x total cattle = (0.29
2 )(2820) = 237.162 c
2 =
å
=
(
O
-
E
)
E
2
(
1430
-
1421.562
1421.562
) 2
+
(
1130
-
1161.276
1161.276
= 0.05 + 0.84 + 2.199 = 3.089
) 2
+
(
260
-
237.162
)
237.162
2 df = 3-1 = 2 critical value = 5.991
Accept the hypothesis; The traits appear to be controlled by a single pair of autosomal genes under random mating.
3.
On the basis of allele-frequency analysis of data from a randomly mating population Snyder (1934) concluded that the ability vs. inability to taste phenylthiocarbamide (PtC) is determined by a single pair of autosomal alleles, of which T for taster is dominant to
T
for nontaster. Of the 3,643 individuals tested in this population, 70% were tasters and 30% were nontasters. Assume the population satisfies the conditions of Hardy-
Weinberg equilibrium. q a) Calculate the frequencies of the alleles T and
T and the frequencies of the genotypes TT, T T and TT .
2 = 30% = 0.30 q = f(
T
) = 0.55 p = f(T) = 0.45 p 2 = f (TT) = 0.20
2 pq = f (T T ) = 0.50 b) Determine the probability of a nontaster child from a taster x taster mating.
Probability (tt) child = P(Tt parent) x P(Tt parent) x P (tt child)
However, we know the parents are not tt, so we must calculate the frequency of Tt parents within the taster population (not the population at large) as follows:
= frequency Tt/ total frequency of tasters (Tt + TT)
= 0.50 / 0.50 + 0.20
= 0.71
So P(tt child from taster parents) = 0.71 x 0.71 x 0.25 = 0.126
4.
In Drosophila melanogaster, Cncn (red vs. cinnabar eyes), Bb ( gray vs. black body, (and Byby (normal vs. blistery wing) are autosomal pairs of alleles.
Samples of three large natural adult populations, each classified for a different pair of traits, are found to have the following genotypes:
Population A
Population B
31 cncn
182 BB
171 Cncn 60 CnCn Total 262
391 Bb 152 bb Total 725
Population C 100 ByBy 372 Byby 40 byby . Total 512
Compare these distributions with those expected for a population at Hardy-
Weinberg equilibrium. Propose a reasonable explanation to account for any differences.
Population A: p = (60 + 85.5)/262 = 0.56; q = 0.44
Expected CnCn = p 2 = 0.31 x 262 = 82 Observed 60
Expected Cncn = 2pq = 0.49 x 262 = 129 Observed 171
Expected cncn = q2 = 0.19 x 262 = 51 Observed 31
Population B: p = 0.52; q = 0.48
Expected BB = 196 Observed BB = 182
Expected Bb = 362 Observed = 391
Expected bb = 167 Observed = 152
Population C: p = 0.56 ; q = 0.44
Expected ByBy = 161
Expected Byby = 252
Expected byby = 99
Observed = 100
Observed = 372
Observed = 40
Without doing Chi-square analysis, all three populations show increased heterozygosity compared to the expected, as well as decreased homozygosity of both genotypes. There may be selection for the heterozygote, or outbreeding
(you’d need to see the same pattern of increased heterozygotes at all genes) or negative assortative mating (if this pattern only existed for one gene and closely linked genes). All three of these situations would give an increase in heterozygotes. As a side note, whatever is going on is much more pronounced in population C.
5.
A yak population is in Hardy-Weinberg equilibrium with allele frequencies p(A) = 0.5 and q(a) = 0.5 for a gene governing color differences. If a new type of predator appears in the area, calculate the equilibrium values of p and q if:
S
AA
= 0.50
S
Aa
= 0.30 s aa
= 0.60 p = S aa
/ S
AA
+S aa
= 0.6/(0.5+0.6) = 0.545 q = 1-0.545 = 0.455
6.
In the population in question 5, calculate the expected new values of p and q after just one generation of selection.
To solve this problem, you must first calculate the fitness values for each genotype (1-S = W).
W
AA
= 0.5
W
Aa
= 0.7
W aa
= 0.4
Then, calculate the mean population fitness using:
W
= p
2
W
AA
+
2 pqW
Aa
+ q
2
W aa
Then calculate the new values for q using the formula: q a
= 1-p a
where: p
A
= p 2 W
AA
+ pqW
Aa
SO:
W W
W
=
.5
2 (0.5)
+
2(.5)(.5)(.7)
+
.5
2 (.4)
=
0.575
p
A
= .5
2 (0.5) + (0.5)(0.5)(0.7)
0.575 0.575
= 0.52 q a
= 1-0.2 = 0.48