Powerpoint
advertisement

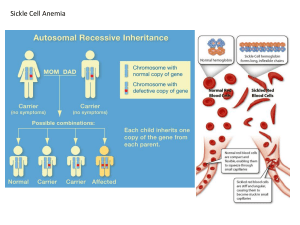
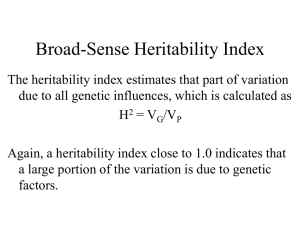
Constant Allele Frequencies Hardy-Weinberg Equilibrium Population An interbreeding group of the same species within a given geographical area Gene pool Population genetics the collection of all alleles in the members of the population the study of the genetics of a population and how the alleles vary with time Gene Flow alleles can move between populations when individuals migrate and mate Allele Frequencies Allelic Frequency # of particular allele total # of alleles in the population Count both chromosomes of each individual Allele frequencies affect the genotype frequencies The frequency of each type of homozygote and heterozygote in the population Phenotype Frequencies Frequency of a trait varies in different populations Table 14.1 Microevolution and Macroevolution Microevolution Genetic change due to changing allelic frequencies in populations Macroevolution The formation of new species Allelic frequencies can change when there is: Nonrandom mating Gene flow Reproductively isolated groups form within or separate from a larger population Mutation e.g. migration Genetic drift Individuals of one genotype are more likely to produce offspring with each other than with those of other genotypes Introduces new alleles into the population Natural selection Individuals with a particular genotype are more likely to produce viable offspring Hardy-Weinberg Equilibrium Developed by mathematicians A condition in which allele frequencies remain constant Used algebra to explain how allele frequencies predicts genotype and phenotype frequencies in equilibrium Hardy-Weinberg Equilibrium p+q=1 All of the allele frequencies together equals 1 or the whole collection of alleles p = allele frequency of one allele (e.g. dominant) q = allele frequency of a second allele (e.g. recessive) p2 + 2pq + q2 = 1 All of the genotype frequencies together equals 1 p2 and q2 =genotype frequencies for each homozygote 2pq = genotype frequency for heterozygotes 2 possible combinations (p egg + q sperm or vice versa) Figure 14.3 Table 14.2 Applying Hardy-Weinberg Equilibrium Used to determine carrier probability Homozygous recessive used to determine frequency of allele in population (phenotype is genotype) Applying Hardy-Weinberg Equilibrium: Cystic Fibrosis Calculating Carrier Frequency for X-linked Traits Figure 14.6 DNA Profiling (a.k.a. DNA Fingerprinting Hardy-Weinberg equilibrium applies to portions of the genome that do not affect phenotype They are not subject to natural selection Short repeated segments that are not protein encoding, distributed all over the genome Detects differences in repeat copy number Calculates probability that certain combinations can occur in two sources of DNA Requires molecular techniques and population studies Preparing DNA for Profiling –Restriction Enzymes Chop up the DNA at specific sequences using “restriction enzymes” Creates RFLPs Restriction fragment length polymorphisms Preparing DNA for Profiling –Running a Gel Run samples on an agarose or polyacrylamide gel DNA has a negative charge so it will travel toward a positive charge Larger fragments will not move as far through the gel DNA Profiling Developed in 1980s Identifies individuals Used in forensics, agriculture, paternity testing, and historical investigations DNA can be obtained from many sources DNA Profiles Figure 14.9 DNA Profiling Types of repeats Variable number tandem repeats (VNTRs) Short tandem repeats (STRs) Shorter than VNTRs Useful if DNA from sample is fragmented or degraded mtDNA Useful if nuclear DNA is highly damaged A Sneeze Identifies Art Thief Table 14.6 Comparing DNA Sequences Figure 14.10 Figure 14.10