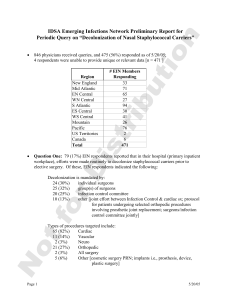
Clinical Infectious Diseases 2007;45:541–547
© 2007 by the Infectious Diseases Society of America. All rights reserved.
1058-4838/2007/4505-0003$15.00
DOI: 10.1086/520663
Mupirocin Resistance in Patients Colonized with
Methicillin‐ Resistant Staphylococcus aureus in a
Surgical Intensive Care Unit
Jeffrey C. Jones,1
Theodore J. Rogers,1
Peter Brookmeyer,1
William Michael Dunne, Jr.,2
Gregory A. Storch,3
Craig M. Coopersmith,4
Victoria J. Fraser,1 and
David K. Warren1
1
Division of Infectious Diseases and Departments of 2Pathology and Immunology,
Pediatrics, and 4Surgery and Anesthesiology, Washington University School of
Medicine, St. Louis, Missouri
3
Background.
Nasal colonization with methicillin‐ resistant Staphylococcus aureus
(MRSA) can be a precursor to serious infection, and decolonization with topical
mupirocin has been studied as a means of preventing clinical infection. Mupirocin
resistance in patients with MRSA has been reported, usually in the context of
widespread mupirocin use.
Methods.
Patients admitted to a surgical intensive care unit (SICU) had nasal swab
cultures for MRSA performed at admission, weekly, and at discharge in an active
surveillance program. Collected MRSA isolates were tested for mupirocin resistance,
and molecular analysis was performed. Clinical data on the characteristics and outcomes
of the patients who stayed in the SICU for >48 h were collected prospectively.
Results.
Of the 302 MRSA isolates available for testing, 13.2% were resistant to
mupirocin, with 8.6% having high‐ level resistance (minimum inhibitory concentration,
512 μg/mL) and 4.6% having low‐ level resistance (minimum inhibitory
concentration, 8–256 μg/mL). Patients admitted to the SICU for >48 h who were
colonized with mupirocin‐ resistant MRSA were more likely to have been admitted to
our hospital during the previous year (
), were older (
), and had higher
in‐ hospital mortality (16% vs. 33%;
), compared with patients colonized with
mupirocin‐ susceptible MRSA. Molecular analysis of the mupirocin‐ resistant isolates
revealed that 72.5% of isolates contained staphylococcal cassette chromosome mec II.
Repetitive sequence polymerase chain reaction typing revealed that high‐ level
mupirocin resistance was present in multiple clonal groups. The rate of mupirocin use
hospital‐ wide during the study period was 6.08 treatment‐ days per 1000
patient‐ days.
Conclusions.
We documented a high rate of mupirocin resistance in MRSA isolates
from SICU patients, despite low levels of in‐ hospital mupirocin use.
Received 18 January 2007; accepted 23 May 2007; electronically published 13 July
2007.
Reprints or correspondence: Dr. Jeffrey C. Jones, Washington University School of
Medicine, Division of Infectious Diseases, 660 S. Euclid Ave., Campus Box 8051, St.
Louis, MO 63110 (jcjones@im.wustl.edu).
Methicillin‐ resistant Staphylococcus aureus (MRSA) is an increasingly common
pathogen associated with both nosocomial and community‐ acquired infections.
Colonization of the anterior nares with S. aureus is common and provides a reservoir for
infection of other sites. Nasal colonization with S. aureus has been linked to
surgical‐ site infection [1], bloodstream infection [2], and ventilator‐ associated
pneumonia [3].
Mupirocin is a topical antibiotic that inhibits bacterial isoleucyl tRNA sythetase,
blocking the formation of isoleucyl tRNA, which in turn impairs bacterial protein
synthesis. Intranasal application of mupirocin is used widely to eliminate S. aureus
colonization and has been studied as a means of preventing health care–associated
staphylococcal infection, such as surgical‐ site infection [4] and bloodstream infection
in patients undergoing hemodialysis [5]. With the recent increase in
community‐ associated MRSA, mupirocin is now being used to decolonize carriers in
an attempt to prevent recurrent infections and to interrupt outbreaks of MRSA‐ related
skin infection [6]. Testing for mupirocin susceptibility is not routinely performed in
most clinical microbiology laboratories.
There are no formally defined breakpoints for mupirocin susceptibility. Since mupirocin
became available in the 1980s, 2 distinct forms of resistance have been described in
association with S. aureus [7]. Low‐ level resistance is mediated by point mutations in
the chromosomal ileS gene. High‐ level resistance to mupirocin is conferred by a novel
gene, mupA, which encodes for an isoleucyl tRNA sythetase enzyme that is not
susceptible to mupirocin and can be acquired through plasmid exchange [8]. Although
local concentrations of mupirocin are high in the areas where it is applied, both high‐
and low‐ level resistance to mupirocin have been associated with decolonization
treatment failure [9, 10]. Widespread use of mupirocin has been linked to increasing
rates of resistance at both an institutional level [11] and a national level [12]. A recent
randomized trial of mupirocin use for MRSA decolonization [13] reported a high rate
(24%) of mupirocin resistance at study enrollment, emphasizing the need to test for
mupirocin resistance prior to implementing routine mupirocin use. We sought to
describe the epidemiology of and risk factors for nasal colonization with
mupirocin‐ resistant MRSA in patients in a surgical, trauma, and burn intensive care
unit (SICU) at an institution where mupirocin use is uncommon.
Methods
Epidemiology and culture collection.
The setting for the study was the 24‐ bed
SICU at Barnes‐ Jewish Hospital, a 1250‐ bed tertiary care hospital in St. Louis,
Missouri. All patients admitted to the SICU from December 2002 through December
2004 had a nasal swab culture performed at hospital admission to detect MRSA
colonization. Surveillance nasal cultures were performed weekly and at hospital
discharge for patients staying >48 h. Patients found to be colonized with MRSA were
placed in isolation with contact precautions. This active surveillance program did not
include a protocol for MRSA decolonization, and decolonization with intranasal
mupirocin was not routinely prescribed by physicians in the SICU. During the period of
the study, patients at Barnes‐ Jewish Hospital were not routinely screened for MRSA or
decolonized prior to elective surgical procedures.
Demographic and clinical data, including comorbid conditions and outcomes, were
collected prospectively for all patients admitted to the SICU for >48 h. Patients were
included in the analysis only once, with their positive nasal culture for MRSA. Data
were analyzed using SPSS, version 12.0 (SPSS). Risk factors for mupirocin resistance
were evaluated using Fisher’s exact test for binomial variables, univariate logistic
regression for variables with
3 categories, and the Mann‐ Whitney U test for
continuous variables. A 2‐ tailed P value <.05 was considered to be statistically
significant. This study was approved by the Washington University Human Studies
Committee (St. Louis, MO).
Microbiology.
Swab specimens were collected from both anterior nares of each
patient, transported and stored at room temperature, and inoculated directly onto
Mannitol salt agar plates (BD Diagnostics). The plates were incubated at 35°C and
examined for growth after 24–48 h. This technique has previously shown similar
sensitivity and specificity to broth culture in our laboratory [14]. Strains that produced
yellow colonies were identified as S. aureus using Gram staining, catalase testing, and
coagulase testing with the Staph Latex agglutination assay (LifeSign). Confirmed S.
aureus isolates were subcultured in Trypase soy broth with 5% sheep blood (BD
Diagnostics) and then on oxacillin screening agar containing 6 μg/mL oxacillin (BD
Diagnostics). Plates were incubated at 35°C for 18–24 h. Strains showing distinct
growth were considered to be methicillin resistant.
Isolates of the first positive culture result for MRSA from each patient were stored in
sterile skim milk at −86°C. Isolates were then subcultured to BBL trypticase soy agar
with 5% sheep blood (BD Diagnostics) and incubated at 35°C for 24 h. A second
subculture was then performed using the same method. Isolated colonies were used to
inoculate sterile saline to visually match a 0.5 McFarland standard and were inoculated
on a Mueller Hinton II plate. A mupirocin Etest (AB Biodisk) strip was applied. After
24 h of incubation at 35°C, the MIC was read. Isolates were classified as susceptible
(MIC, <8 μg/mL), low‐ level resistant (MIC, 8–256 μg/mL), or high‐ level resistant
(MIC,
512 μg/mL).
Molecular analysis.
All mupirocin‐ resistant isolates and 7 randomly selected
mupirocin‐ susceptible isolates from the same period were grown overnight in
trypticase soy broth and then centrifuged. DNA was extracted from the pellets using the
method described by Kalia et al. [15] in combination with a QIAamp DNA extraction
kit (Qiagen). Repetitive sequence PCR using RW3A primers was performed using the
method described by Del Vecchio et al. [16]. Molecular Analyst Software (Bio‐ Rad
Laboratories) was used to determine the relatedness of the strains.
PCR amplification was performed on each isolate for detection of mecA, lukS‐ PV, and
lukF‐ PV genes, as previously described [17, 18]. The staphylococcal cassette
chromosome (SCC) mec element was typed in each isolate using the real‐ time
multiplex PCR method described by Francois et al. [19].
Antibiotic usage.
The inpatient pharmacy records of each patient who was positive
for MRSA for the current hospital admission were reviewed for documentation of
mupirocin use. An additional manual chart review of all admissions to our hospital
during the previous year was performed for the 40 patients found to have
mupirocin‐ resistant MRSA. Outpatient pharmacy records were not available for
review. The amounts of mupirocin and other commonly prescribed antibiotics used
hospital‐ wide during each month of the study period were determined from hospital
pharmacy records.
Results
The distribution of the study population is summarized in figure 1. During the study
period, 338 patients (13.6%) admitted to the SICU had a nasal swab culture result
positive for MRSA. Thirty‐ six of the samples from these patients were not saved and
were unavailable for mupirocin susceptibility testing. Forty (13.2%) of the 302 isolates
available were resistant to mupirocin (14 [4.6%] with low‐ level resistance and 26
[8.6%] with high‐ level resistance).
Figure
1.
(27 KB)
Figure
1.
Distribution of the study population. MRSA, methicillin‐ resistant
Staphylococcus aureus; (+)MRSA, nasal swab specimen tested positive for MRSA;
(−)MRSA, nasal swab specimen tested negative for MRSA; SICU, surgical intensive
care unit. aThirty‐ six MRSA isolates were not stored and were unavailable for further
testing.
The characteristics of the 225 patients who had nasal swab culture results positive for
MRSA and who stayed >48 h in the SICU are presented in table 1. Thirty (13.3%) of
these patients had mupirocin‐ resistant MRSA. A comparison of the patients with
mupirocin‐ susceptible MRSA with patients with mupirocin‐ resistant MRSA is
presented in table 1. Patients colonized with mupirocin‐ resistant MRSA were
significantly older (mean age, 70.9 years vs. 61.4 years;
) and were more likely to
have been admitted to Barnes‐ Jewish Hospital in the previous 12 months than were
patients with mupirocin‐ susceptible MRSA (
). In‐ hospital mortality was higher
for patients colonized with mupirocin‐ resistant MRSA than for patients colonized with
mupirocin‐ susceptible MRSA (33% vs. 16%;
). Patients with mupirocin‐ resistant
and mupirocin‐ susceptible MRSA were equally likely to have acquired MRSA in the
SICU (37% vs. 37%;
).
Table
1.
Comparison of patients admitted to the surgical intensive care unit
(SICU) for >48 h who were colonized with mupirocin‐ resistant methicillin‐ resistant
Staphylococcus aureus (MRSA) with patients colonized with mupirocin‐ susceptible
MRSA.
Results of molecular typing of the 40 mupirocin‐ resistant MRSA isolates and selected
susceptible isolates are presented in figure 2. Twenty‐ nine of the mupirocin‐ resistant
MRSA isolates (72.5%) contained SCCmec type II, 5 (12.5%) were positive for
SCCmec type IV, and 6 tested negative for both. No isolate tested positive for lukS‐ PV
or lukF‐ PV, which are genes encoding for Panton‐ Valentine leukocidin toxin. Cluster
analysis of the repetitive sequence PCR results of the 40 mupirocin‐ resistant strains
and 7 mupirocin‐ susceptible MRSA strains revealed 1 large cluster consisting of 29
SCCmec II–positive isolates. This cluster contained 13 high‐ level and 12 low‐ level
mupirocin‐ resistant strains, as well as 4 mupirocin‐ susceptible isolates. Two smaller
clusters containing more SCCmec IV–positive isolates were also identified and
contained a greater proportion of high‐ level resistant isolates. The 6 strains that tested
negative for both SCCmec II and IV were submitted to a reference laboratory for further
testing. One was positive for SCCmec III, and the other 5 were untypable with standard
primers for SCCmec but were phenotypically methicillin resistant and mecA positive by
direct PCR primer assay for the gene.
Figure
2.
(101 KB)
Figure
2.
Cluster analysis of PCR products from 40 mupirocin‐ resistant
methicillin‐ resistant Staphylococcus aureus (MRSA) strains and 7
mupirocin‐ susceptible MRSA strains using repetitive‐ sequence PCR with RW3A
primers. Of the 47 isolates tested, 7 were susceptible, 26 were high‐ level resistant, and
14 were low‐ level resistant. 4, SCCmec IV; H, high‐ level mupirocin resistance (MIC,
512 μg/mL); L, low‐ level resistance (MIC, 8–256 μg/mL); Mu, mupirocin; −,
negative for both SCCmec II and IV (see Results); S, mupirocin susceptible (MIC, <8
μg/mL); 2, SCCmec II.
The rate of mupirocin use hospital‐ wide during the study period was 6.08
treatment‐ days per 1000 patient‐ days. In the SICU, the rate of mupirocin use was 2.29
treatment‐ days per 1000 patient‐ days during the study period. The amounts of
vancomycin, cefepime, and all fluoroquinolones dispensed hospital‐ wide during the
study period were 132.5, 69.98, and 95.06 defined daily doses per 1000 patient‐ days,
respectively.
Two patients whose culture results were positive for MRSA had received mupirocin
during the current hospital admission and prior to their first positive nasal culture result.
One patient had mupirocin‐ susceptible MRSA, and 1 patient had high‐ level
mupirocin‐ resistant MRSA. Both had received mupirocin topically for skin wounds.
None of the patients with mupirocin‐ resistant MRSA received mupirocin at our
hospital during the year prior to the current admission.
Discussion
Previous reports of mupirocin resistance in patients with S. aureus have primarily been
from institutions with high levels of resistance in the context of widespread mupirocin
use, with rates of resistance ranging from 11.3% to 65% [11, 20–23]. Microbiologic
studies without detailed epidemiologic data have reported lower rates of mupirocin
resistance [24–26]. The rate of mupirocin resistance in our study population (13.3%) is
on the higher end of the range of rates of resistance (4.6%–17.8%) from MRSA clinical
isolates in the SENTRY study in the United States. To our knowledge, the rate in our
study is the highest rate of resistance reported from a single institution in the absence of
widespread, routine use of mupirocin.
The majority of the mupirocin‐ resistant MRSA isolates in our study population were
positive for SCCmec II, which is consistent with the traditional health care–associated
MRSA, rather than community‐ associated MRSA. High‐ and low‐ level mupirocin
resistance was found in all clusters identified by repetitive sequence PCR, suggesting
that resistance was not because of dissemination of a single clone. This is consistent
with a recent report showing that high‐ level mupirocin resistance was distributed
among different clones of MRSA in a hospital [27]. This pattern likely results from the
acquisition of plasmids bearing resistance genes either from other S. aureus strains or
from coagulase‐ negative Staphylococcus species, a phenomenon that has been
documented in vitro [28] and, recently, in vivo [29].
Patients with mupirocin‐ resistant MRSA in our study were not more likely to have
acquired colonization while in the SICU. However, the epidemiologic data do support
the hypothesis that mupirocin‐ resistant MRSA colonization among patients in this
study was acquired in the health care environment. Patients colonized with
mupirocin‐ resistant MRSA were significantly more likely to have been admitted to our
hospital in the previous year than were patients with mupirocin‐ susceptible MRSA.
Patients with mupirocin‐ resistant MRSA were also significantly older than patients
with mupirocin‐ susceptible MRSA, which along with the higher in‐ hospital mortality
in this group, seems to indicate a sicker population with more contact with the health
care system prior to admission.
We have documented that the patients in our study had limited exposure to mupirocin
during the current hospitalization, and that there was no use as inpatients at our hospital
during the prior year by the patients with mupirocin‐ resistant strains. A limitation of
the study is the lack of data regarding outpatient use of mupirocin. We cannot rule out
the possibility that patients were receiving treatment with mupirocin as outpatients,
leading to higher rates of resistance. However, we can state that it was not the policy or
routine practice of our institution to screen patients for MRSA preoperatively or to
attempt to decolonize patients known to be MRSA carriers.
Our study reveals a high rate of mupirocin resistance, including high‐ level resistance,
despite low mupirocin use at our institution. This could provide a substrate for more
widespread resistance if selective pressure was applied by increasing mupirocin use.
Testing for mupirocin resistance is not routine at most institutions. The significant rate
of resistance at our hospital, where use of mupirocin is low, highlights the need for
baseline testing and subsequent monitoring for mupirocin resistance before
implementing infection‐ control strategies that rely heavily on mupirocin for MRSA
decolonization.
Acknowledgments
We thank Joan Hoppe‐ Bauer, for her assistance in storing and managing the isolates;
Rebecca Guth, for her assistance with the statistical analysis; and Linda McDougal and
Dr. Brandi Limbago (Centers for Disease Control and Prevention, Atlanta, GA), for
their assistance in further characterizing the nontypable methicillin‐ resistant
Staphylococcus aureus isolates.
Financial support.
Centers for Disease Control and Prevention (1U1CI000033301)
and National Institutes of Health/National Institute of Allergies and Infectious Diseases
(5K23AI050 585‐ 02 to D.K.W.)
Potential conflicts of interest.
References
All authors: no conflicts.
1.
Wenzel RP, Perl TM. The significance of nasal carriage of
Staphylococcus aureus and the incidence of postoperative wound infection. J
Hosp Infect 1995;
31:13–24.
First citation in article, CrossRef, PubMed
2.
von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as
a source of Staphylococcus aureus bacteremia. N Engl J Med 2001;
6.
344:11–
First citation in article, CrossRef, PubMed
3.
Corne P, Marchandin H, Jonquet O, Campos J, Banuls AL. Molecular
evidence that nasal carriage of Staphylococcus aureus plays a role in respiratory
tract infections of critically ill patients. J Clin Microbiol 2005;
43:3491–3.
First citation in article, CrossRef, PubMed
4.
Perl TM, Cullen JJ, Wenzel RP, et al. Intranasal mupirocin to prevent
postoperative Staphylococcus aureus infections. N Engl J Med 2002;
346:1871–7.
First citation in article, CrossRef, PubMed
5.
Boelaert JR, Van Landuyt HW, Godard CA, et al. Nasal mupirocin
ointment decreases the incidence of Staphylococcus aureus bacteraemias in
haemodialysis patients. Nephrol Dial Transplant 1993;
8:235–9.
First citation in article, PubMed
6.
Rihn JA, Posfay‐ Barbe K, Harner CD, et al. Community‐ acquired
methicillin‐ resistant Staphylococcus aureus outbreak in a local high school
football team unsuccessful interventions. Pediatr Infect Dis J 2005;
3.
First citation in article, CrossRef, PubMed
24:841–
7.
Cookson BD. The emergence of mupirocin resistance: a challenge to
infection control and antibiotic prescribing practice. J Antimicrob Chemother
1998;
41:11–8.
First citation in article, CrossRef, PubMed
8.
Hodgson JE, Curnock SP, Dyke KG, Morris R, Sylvester DR, Gross MS.
Molecular characterization of the gene encoding high‐ level mupirocin
resistance in Staphylococcus aureus J2870. Antimicrob Agents Chemother
1994;
38:1205–8.
First citation in article, PubMed
9.
Walker ES, Vasquez JE, Dula R, Bullock H, Sarubbi FA.
Mupirocin‐ resistant, methicillin‐ resistant Staphylococcus aureus: does
mupirocin remain effective? Infect Control Hosp Epidemiol 2003;
6.
24:342–
First citation in article, Abstract, PubMed
10.
Harbarth S, Dharan S, Liassine N, Herrault P, Auckenthaler R, Pittet D.
Randomized, placebo‐ controlled, double‐ blind trial to evaluate the efficacy of
mupirocin for eradicating carriage of methicillin‐ resistant Staphylococcus
aureus. Antimicrob Agents Chemother 1999;
43:1412–6.
First citation in article, PubMed
11.
Miller MA, Dascal A, Portnoy J, Mendelson J. Development of
mupirocin resistance among methicillin‐ resistant Staphylococcus aureus after
widespread use of nasal mupirocin ointment. Infect Control Hosp Epidemiol
1996;
17:811–3.
First citation in article, PubMed
12.
Upton A, Lang S, Heffernan H. Mupirocin and Staphylococcus aureus:
a recent paradigm of emerging antibiotic resistance. J Antimicrob Chemother
2003;
51:613–7.
First citation in article, CrossRef, PubMed
13.
Simor AE, Phillips E, McGeer A, et al. Randomized controlled trial of
chlorhexidine gluconate for washing, intranasal mupirocin, and rifampin and
doxycycline versus no treatment for the eradication of methicillin‐ resistant
Staphylococcus aureus colonization. Clin Infect Dis 2007;
44:178–85.
First citation in article, Abstract, PubMed
14.
Warren DK, Liao RS, Merz LR, Eveland M, Dunne WM Jr. Detection
of methicillin‐ resistant Staphylococcus aureus directly from nasal swab
specimens by a real‐ time PCR assay. J Clin Microbiol 2004;
42:5578–81.
First citation in article, CrossRef, PubMed
15.
Kalia A, Rattan A, Chopra P. A method for extraction of high‐ quality
and high‐ quantity genomic DNA generally applicable to pathogenic bacteria.
Anal Biochem 1999;
275:1–5.
First citation in article, CrossRef, PubMed
16.
Del Vecchio VG, Petroziello JM, Gress MJ, et al. Molecular genotyping
of methicillin‐ resistant Staphylococcus aureus via fluorophore‐ enhanced
repetitive‐ sequence PCR. J Clin Microbiol 1995;
33:2141–4.
First citation in article, PubMed
17.
Jarraud S, Mougel C, Thioulouse J, et al. Relationships between
Staphylococcus aureus genetic background, virulence factors, agr groups
(alleles), and human disease. Infect Immun 2002;
70:631–41.
First citation in article, CrossRef, PubMed
18.
Strommenger B, Kettlitz C, Werner G, Witte W. Multiplex PCR assay
for simultaneous detection of nine clinically relevant antibiotic resistance genes
in Staphylococcus aureus. J Clin Microbiol 2003;
First citation in article, CrossRef, PubMed
41:4089–94.
19.
Francois P, Pittet D, Bento M, et al. Rapid detection of
methicillin‐ resistant Staphylococcus aureus directly from sterile or nonsterile
clinical samples by a new molecular assay. J Clin Microbiol 2003;
60.
41:254–
First citation in article, CrossRef, PubMed
20.
Vivoni AM, Santos KR, de‐ Oliveira MP, et al. Mupirocin for
controlling methicillin‐ resistant Staphylococcus aureus: lessons from a decade
of use at a university hospital. Infect Control Hosp Epidemiol 2005;
26:662–7.
First citation in article, Abstract, PubMed
21.
Vasquez JE, Walker ES, Franzus BW, Overbay BK, Reagan DR,
Sarubbi FA. The epidemiology of mupirocin resistance among
methicillin‐ resistant Staphylococcus aureus at a Veterans' Affairs hospital.
Infect Control Hosp Epidemiol 2000;
21:459–64.
First citation in article, Abstract, PubMed
22.
Leski TA, Gniadkowski M, Skoczynska A, Stefaniuk E, Trzcinski K,
Hryniewicz W. Outbreak of mupirocin‐ resistant staphylococci in a hospital in
Warsaw, Poland, due to plasmid transmission and clonal spread of several
strains. J Clin Microbiol 1999;
37:2781–8.
First citation in article, PubMed
23.
Walker ES, Levy F, Shorman M, David G, Abdalla J, Sarubbi FA. A
decline in mupirocin resistance in methicillin‐ resistant Staphylococcus aureus
accompanied administrative control of prescriptions. J Clin Microbiol
2004;
42:2792–5.
First citation in article, CrossRef, PubMed
24.
Deshpande LM, Fix AM, Pfaller MA, Jones RN. Emerging elevated
mupirocin resistance rates among staphylococcal isolates in the SENTRY
Antimicrobial Surveillance Program (2000): correlations of results from disk
diffusion, Etest and reference dilution methods. Diagn Microbiol Infect Dis
2002;
42:283–90.
First citation in article, CrossRef, PubMed
25.
Schmitz FJ, Lindenlauf E, Hofmann B, et al. The prevalence of low‐
and high‐ level mupirocin resistance in staphylococci from 19 European
hospitals. J Antimicrob Chemother 1998;
42:489–95.
First citation in article, CrossRef, PubMed
26.
Kresken M, Hafner D, Schmitz FJ, Wichelhaus TA. Prevalence of
mupirocin resistance in clinical isolates of Staphylococcus aureus and
Staphylococcus epidermidis: results of the Antimicrobial Resistance
Surveillance Study of the Paul‐ Ehrlich‐ Society for Chemotherapy, 2001. Int J
Antimicrob Agents 2004;
23:577–81.
First citation in article, CrossRef, PubMed
27.
Perez‐ Roth E, Lopez‐ Aguilar C, coba‐ Florez J, Mendez‐ Alvarez S.
High‐ level mupirocin resistance within methicillin‐ resistant Staphylococcus
aureus pandemic lineages. Antimicrob Agents Chemother 2006;
11.
50:3207–
First citation in article, CrossRef, PubMed
28.
Bastos MC, Mondino PJ, Azevedo ML, Santos KR,
Giambiagi‐ deMarval M. Molecular characterization and transfer among
Staphylococcus strains of a plasmid conferring high‐ level resistance to
mupirocin. Eur J Clin Microbiol Infect Dis 1999;
18:393–8.
First citation in article, CrossRef, PubMed
29.
Hurdle JG, O’Neill AJ, Mody L, Chopra I, Bradley SF. In vivo transfer
of high‐ level mupirocin resistance from Staphylococcus epidermidis to
methicillin‐ resistant Staphylococcus aureus associated with failure of
mupirocin prophylaxis. J Antimicrob Chemother 2005;
First citation in article, CrossRef, PubMed
56:1166–8.
Cited by
Kanokporn Mongkolrattanothai, MD; Peggy Mankin, BS, MA; Venkedesh Raju, MD;
Barry Gray, MD. (2008) Surveillance for Mupirocin Resistance Among MethicillinResistant Staphylococcus aureus Clinical Isolates • . Infection Control and Hospital
Epidemiology 29:10, 993-994
Online publication date: 1-Oct-2008.
Citation-Full Text-PDF Version (308 kB)
X. Malaviolle, C. Nonhoff, O. Denis, S. Rottiers, M. J. Struelens. (2008) Evaluation of
disc diffusion methods and Vitek 2 automated system for testing susceptibility to
mupirocin in Staphylococcus aureus. Journal of Antimicrobial Chemotherapy 62:5,
1018-1023
Online publication date: 18-Aug-2008.
CrossRef
H. Humphreys. (2008) Can we do better in controlling and preventing methicillinresistant Staphylococcus aureus (MRSA) in the intensive care unit (ICU)?. European
Journal of Clinical Microbiology & Infectious Diseases 27:6, 409-413
Online publication date: 1-Jul-2008.
CrossRef
Carlo Gelmetti. (2008) Local antibiotics in dermatology. Dermatologic Therapy 21:3,
187-195
Online publication date: 1-Jun-2008.
CrossRef
Itzhak Brook. (2008) Clinical trials report. Current Infectious Disease Reports 10:3,
179-181
Online publication date: 1-Jun-2008.
CrossRef
Michael R Jacobs. (2008) Retapamulin: a semisynthetic pleuromutilin compound for
topical treatment of skin infections in adults and children. Future Microbiology 2:6,
591-600
Online publication date: 1-Jan-2008.
CrossRef