Chapter 11
advertisement
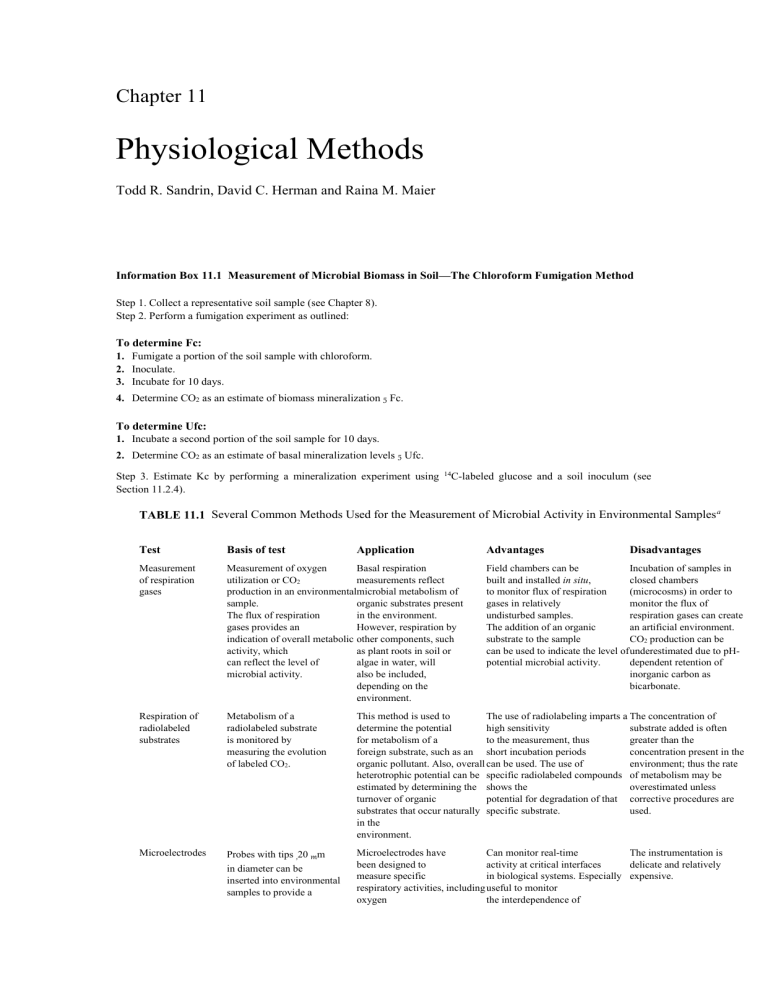
Chapter 11 Physiological Methods Todd R. Sandrin, David C. Herman and Raina M. Maier Information Box 11.1 Measurement of Microbial Biomass in Soil—The Chloroform Fumigation Method Step 1. Collect a representative soil sample (see Chapter 8). Step 2. Perform a fumigation experiment as outlined: To determine Fc: 1. Fumigate a portion of the soil sample with chloroform. 2. Inoculate. 3. Incubate for 10 days. 4. Determine CO2 as an estimate of biomass mineralization 5 Fc. To determine Ufc: 1. Incubate a second portion of the soil sample for 10 days. 2. Determine CO2 as an estimate of basal mineralization levels 5 Ufc. Step 3. Estimate Kc by performing a mineralization experiment using Section 11.2.4). 14 C-labeled glucose and a soil inoculum (see TABLE 11.1 Several Common Methods Used for the Measurement of Microbial Activity in Environmental Samples a Test Basis of test Measurement of respiration gases Measurement of oxygen Basal respiration utilization or CO2 measurements reflect production in an environmentalmicrobial metabolism of sample. organic substrates present The flux of respiration in the environment. gases provides an However, respiration by indication of overall metabolic other components, such activity, which as plant roots in soil or can reflect the level of algae in water, will microbial activity. also be included, depending on the environment. Respiration of radiolabeled substrates Metabolism of a radiolabeled substrate is monitored by measuring the evolution of labeled CO2. This method is used to The use of radiolabeling imparts a The concentration of determine the potential high sensitivity substrate added is often for metabolism of a to the measurement, thus greater than the foreign substrate, such as an short incubation periods concentration present in the organic pollutant. Also, overall can be used. The use of environment; thus the rate heterotrophic potential can be specific radiolabeled compounds of metabolism may be estimated by determining the shows the overestimated unless turnover of organic potential for degradation of that corrective procedures are substrates that occur naturally specific substrate. used. in the environment. Microelectrodes Probes with tips ,20 mm Microelectrodes have Can monitor real-time The instrumentation is been designed to activity at critical interfaces delicate and relatively measure specific in biological systems. Especially expensive. respiratory activities, including useful to monitor oxygen the interdependence of in diameter can be inserted into environmental samples to provide a Application Advantages Disadvantages Field chambers can be Incubation of samples in built and installed in situ, closed chambers to monitor flux of respiration (microcosms) in order to gases in relatively monitor the flux of undisturbed samples. respiration gases can create The addition of an organic an artificial environment. substrate to the sample CO2 production can be can be used to indicate the level of underestimated due to pHpotential microbial activity. dependent retention of inorganic carbon as bicarbonate. Incorporation of radiolabeled thymidine into cellular DNA Adenylate energy charge (AEC) continuous monitoring of activity. utilization and nitrate respiration. aerobic and anaerobic processes in the environment. Microorganisms will scavenge The rate of DNA When using a short Not all bacteria will DNA precursors, such as synthesis provides a incubation time, thymidine incorporate exogenously thymidine, from their reasonable estimate of incorporation is thought to supplied thymidine into environment. By radiolabeling the rate of cell division, measure bacterial DNA DNA. Also, estimating thymidine, thus providing an production because the rate of microbial activity requires the rate of incorporation estimate of microbial biomass bacterial incorporation of the development of a into DNA can be production. thymidine is thought to be much conversion factor relating measured. faster than for other organisms thymidine incorporation to which may be present in biomass production. environmental samples. AEC is a weighted ratio of ATP to total adenylates. ATP is quantified using a luciferin–luciferase substrate– enzyme system. AEC values reflect a continuum between an active microbial community (AEC . 0.8) and a community with a high proportion of dead or moribund cells (AEC , 0.4). AEC can establish the AEC is not necessarily a presence of a metabolically active direct measure of microbial community without activity because adenylates the need to incubate the sample or are present in all living add a surrogate substrate. organisms and can also be released by decaying cells. Dehydrogenase assay This assay measures The rate of reduction of Actively respiring The amount of tetrazolium the rate of oxidation–reduction tetrazolium salts reflects the microorganisms can be visualized salts reduced depends on reactions (electron transport overall activity by all respiring microscopically many factors including chain activity) by monitoring microorganisms. by the deposition of sample incubation the reduction of a tetrazolium pigmented tetrazolium salt conditions. Direct salt by reduction products within the cell. comparison of activity actively respiring The assay has a high sensitivity, between samples can be microorganisms. and can be used to measure made only when the assay activity in low productivity is performed under environments. identical conditions. Hydrolysis of fluorescein diacetate Hydrolysis of fluorescein diacetate is performed by a variety of enzymes including esterases, proteases, and lipases. The assay measures enzymatic Fluorescein diacetate hydrolysis activity, thus providing an produces a highly fluorescent estimate product, fluorescein, which is of total microbial activity easily detected. in an environmental sample. Enzymes involved in hydrolysis can be intracellular or extracellular. Stable isotope probing (SIP) An isotopically labeled (e.g., 13C) substrate is added to a sample. Analysis of 13C distribution into DNA (DNA-SIP) or cell lipids (PFLA-SIP) provides information about the active populations in the community. This technique identifies the populations within a community that are actively metabolizing the labeled substrate added. Links phylogeny with function even if the functional populations cannot be cultured. Can be used to measure carbon flow through trophic levels in the environment. PFLA-SIP is more sensitive in terms of numbers but gives only very general taxonomic information. DNA-SIP is less sensitive in terms of number but gives precise phylogenetic information. SIP is technically difficult and not yet standardized. Microarray A matrix of immobilized Allows high throughput nucleic acid probes is screening of gene expression used to query the nucleic acid in a sample. makeup in a sample. Allows direct and rapid inquiry of how microorganisms respond to and interact with their environment. Cost prohibitive in many cases although technological advances are decreasing costs associated with microarray fabrication. Data interpretation is also problematic. Proteomics Analysis of the proteins expressed by a microorganism under a prescribed set of environmental conditions. a Allows identification of Provides the best Analysis is expensive, proteins that are differentially information available describing a difficult, and timeexpressed and, thus, likely microbial response to a stimulus. consuming. The technique important in microbial has not yet been made userresponses to environmental friendly. conditions. This table is meant to highlight advantages and disadvantages of these techniques, and should not be considered as comprehensive. TABLE 11.2 General and Specific Enzyme Assays That Can Be Used to Measure Microbial Activity Enzyme Substrate Description of assay Dehydrogenase Triphenyltetrazolium Dehydrogenases convert triphenyltetrazolium chloride to triphenylformazan; the triphenylformazan is extracted with methanol and quantitated spectrophotometrically. Phosphatase p-Nitrophenol phosphate Phosphatases convert the p-nitrophenol phosphate to p-nitrophenol, which is extracted in aqueous solution and quantitated spectrophotometrically. Protease Gelatin Gelatin hydrolysis, as an example of proteolytic activity, can be measured by the determination of residual protein. Amylase Starch The amount of residual starch is quantitated spectrophotometrically by the intensity of the blue color resulting from its reaction with iodine. Chitinase Chitin Production of reducing sugars is measured using anthrone reagent. Cellulase Cellulose, carboxymethylcellulose Production of reducing sugars is measured using anthrone reagent. Cellulases alter the viscosity of carboxymethylcellulose, a quantity that can be measured. Nitrogenase Acetylene Nitrogenase, besides reducing dinitrogen gas (N2) to ammonia (NH3), is also capable of reducing acetylene (C2H2) to ethylene (C2H4); the rate of formation of ethylene can be monitored using a gas chromatograph, and the rate of nitrogen fixation can be calculated using an appropriate conversion factor. Nitrate reductase Nitrate Dissimilatory nitrate reductase can be assayed by the disappearance of nitrate or by measuring with a gas chromatograph the evolution of denitrification products, such as nitrogen gas and nitrous oxide, from samples; denitrification can be blocked at the nitrous oxide level by the addition of acetylene, permitting a simpler assay procedure. From Atlas and Bartha, 1993.











