Abstract
advertisement

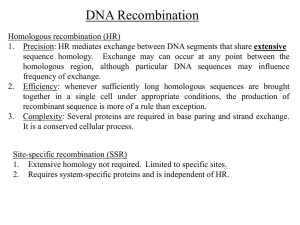
converged? stop : otherwise back to (1) Bates-Paul (P0096) - 72 predictions: 72 3D Comparative Modelling By In Silico Recombination of Templates, Alignments and Models This is a standard genetic algorithm with two genetic operators (recombination and mutation) and a fitness function acting as an artificial selection agent. We will now briefly describe each step in the protocol. Bruno Contreras-Moreira, Paul W. Fitzjohn, Marc Offman, Graham R. Smith and Paul A. Bates Initial population of models. Initially, our server Domain Fishing [3] (www.bmm.icnet.uk/servers/ 3djigsaw/dom_fish) was used to define protein domains within each target sequence and to find suitable modelling templates. Resulting alignments were inspected and corrected if suspected to be incorrect. If reasonable alternative alignments could be found they too were added to the pool. When possible, only alignments with bit-scores (average pssm-logodds+secondary structure agreement/residue) around 2 were selected. In several cases annotations from the templates or their corresponding PFAM families were used to check the correctness of the alignment in active/binding sites. Usually several models were built using the same template changing parts in the alignment. Models from these alignments were built using our server 3D-JIGSAW [4] (www.bmm.icnet.uk/servers/3djigsaw). Additional models were obtained from the CAFASP3 server after inspection of the alignments to gain extra variability in sequence alignments, templates used and exposed loops. These models were taken from different sources, including FAMS (physchem.pharm.kitasatou.ac.jp/FAMS), Pmodeller (www.sbc.su.se/~arne/pcons) and EsyPred3D (www.fundp.ac.be/urbm/bioinfo/esypred). Models were inspected and missing parts, typically loops, added using in-house software before going to the next step. In essence, this software explores phi/psi space to allow a peptide (the missing loop) to connect a gap in a protein fold. Biomolecular Modelling Laboratory Cancer Research UK - London Research Institute paul.bates@cancer.org.uk After the CASP4 assessment it was concluded that template selection and sequence alignment remain the main problems awaiting solution in the field of comparative modelling [1]. Models were rarely found to be closer to the experimental structures than the optimal template and often manual intervention only marginally mproved their quality. Similar problems were found in the fold recognition category [2,4], suggesting that the same approach may be applied in the search for possible solutions in both fields. During CASP5 our group has tested a novel procedure to tackle these problems. This new method was used to generate models for all 67 targets, with roughly half of them classified as fold recognition targets by the CAFASP3 meta-server (www.cs.bgu.ac.il/~dfischer/CAFASP3). This procedure is named in silico protein recombination, as it is a computational implementation of genetic recombination, a well known mechanism for generating population variability, but at the protein level. For each CASP5 target a population of models was generated from a variety of templates and sequence alignments. Care was taken to assure that models had similar length and were complete, adding missing loops when necessary and smoothing their phi/psi geometry to permit later energy calculations and minimizations. The algorithm can be outlined as: 1. Growing the population by recombination and mutation. The initial population was grown by randomly selecting pairs of protein models and applying one of the two possible operators. In the case of recombination, the models were superimposed based on their sequence alignment and a crossover point drawn. Crossover was not permitted inside secondary structure elements. The resulting recombinant model inherits the N-terminus from one parent and the C-terminus from the other. In mutation events (occurring with frequency 1r, where r is the recombination probability) a new protein model was obtained initial population of models (1) grow population: r recombination + (1-r) mutation (2) select best proportion according to fitness A-1 by simply averaging its parents' coordinates after superimposition. In many cases this process obtained distorted side-chain conformations. 2. Sippl M.J., Lackner P., Domingues F.S., Prlic A., Malik R., Andreeva A. 2. Selecting the best proportion. Fitness function. The whole idea of the algorithm is that it should be possible to obtain optimized mosaic models by shuffling them in a rational way. The key point in this approach is thus the choice of an appropriate fitness function. After some benchmarking experiments (unpublished results) we chose a function that calculates a free energy estimate based on two terms: protein contact pair-potentials and sidechain solvation energies estimated from their solvent accessible area. This function seems to yield a consistent measure of protein structural quality. When each population reaches the upper limit (between 2 and 4 times its initial size), this energy function is used to rank its members. Only the worst 25% of the population is discarded at this point, to assure that quality models are not lost prematurely. 3. and Wiederstein M.(2001) Assessment of the CASP4 Fold Recognition Category. Protein suppl 5, 55-67. Contreras-Moreira B. and Bates P.A. (2002) Domain Fishing: a first step in protein comparative modelling. Bioinformatics 18, 1141-1142. 4. Bates P.A., Kelley L.A., MacCallum R.M. and Sternberg M.J.E. (2001) Enhancement of Protein Modelling by Human Intervention in Applying the Automatic Programs 3DJIGSAW and 3D-PSSM. Proteins suppl 5, 39-46. (www.bmm.icnet.uk/servers/3djigsaw) 3. Convergence criterion and final refinements. When the population has converged to similar energies, there is no room for further generation of variability and the evolution process stops. At this point the final population is inspected. In most cases this consists of several representations of the same protein conformation with average backbone deviations in the order of 0.1Å. One of these representatives is then taken as the final model, which is carefully inspected to detect unfavorable peptide conformations and a final energy minimization using the CHARMM22 force field is performed. This procedure is able to fix distorted side-chains. At this point we have a CASP5 unrefined model. In addition, for targets T0134, T0165, T0177 and T0185 we tested a further refinement step consisting of running an all-atom, molecular dynamics simulation inside a water box, with neutral total charge for around 0.5ns. For these simulations we used the GROMACS package (www.gromacs.org) and the OPLSAA force field. Snapshots taken from the trajectory were clustered according to average backbone deviations and one conformation from the most populated cluster was selected. After a few rounds of CHARMM22 energy minimization, it was submitted as a refined model. Insufficient computer resources prevented us from refining all targets. 1. Tramontano A., Leplae R. and Morea V. (2001) Analysis and Assessment of Comparative Modeling Predictions in CASP4.. Proteins suppl 5, 22-38 A-2