Lecture 8
advertisement

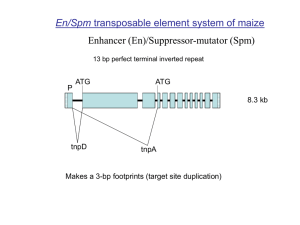
DNA Recombination Homologous recombination (HR) 1. Precision: HR mediates exchange between DNA segments that share extensive sequence homology. Exchange may can occur at any point between the homologous region, although particular DNA sequences may influence frequency of exchange. 2. Efficiency: whenever sufficiently long homologous sequences are brought together in a single cell under appropriate conditions, the production of recombinant sequence is more of a rule than exception. 3. Complexity: Several proteins are required in base paring and strand exchange. It is a conserved cellular process. Site-specific recombination (SSR) 1. Extensive homology not required. Limited to specific sites. 2. Requires system-specific proteins and is independent of HR. Site-Specific Recombination (SSR) 1. Conservative SSR (CSSR): i) Recombination occurs at specific sites, within short sequence identity. ii) strand exchange occurs by precise breakage and joining events. iii) no synthesis or loss of DNA sequences occur. iv) recombination is reciprocal. 2. Non-conservative or Transpositional: i) no homology between the recombination sites ii) DNA synthesis accompanies breakage and joining events. iii) recombination is non-reciprocal. Biological consequences of recombination Integration/Excision system: lambda (λ) system Lysogeny attP + attB IHF Integrase (Int) attL + attR Prophage induction IHF attL + attR Integrase (Int) attP + attB Xis (phage encoded) Other phages such as phi80, P2, P4 and P22 also employ CSSR integration/excision system Biological consequences of recombination Plasmid segregation: Stable plasmid maintenance requires that plasmid copies efficiently distribute into daughter cells at cell division. High copy plasmids are randomly distributed. Low copy plasmids have to use specialized partitioning system. Both mechanisms require that the presence of multimeric plasmids, which can be generated by homologous recombination. These multimers must be minimized to monomers prior to partitioning. P1 phage uses site-specific recombination system, Cre-lox, to resolve multimers into monomers. Single protein bearing recombinase activity, Cre, is required for the reversible recombination reaction. lox Cre lox Cre-lox P1 phage has circular genome but linear genetic map. Recombinase: Cre DNA recombination substrate: loxP 34 bp (13 bp inverted repeats flank 8 bp asymmetric sequence) ATAACTTCGTATA GCATACAT TATACGAAGTTAT TATTGAAGCATAT CGTATGTA ATATGCTTCAATA LoxP sequence 13-8-13 Biological consequences of recombination Inversion system (The FLP-FRT system): The 2-micron circular plasmid often found in yeast encodes a CSSR system that can invert a large segment of plasmid. The inversion changes the orientation of replicative forks within the plasmid and thereby promotes amplification to high copy number. FRT FLP Rep fork Rep fork Rep fork Rep fork FLP-FRT Recombinase: FLP Recombination site: FRT 13-13-8-13 13-8-13 FRT: original FRT R-RS Recombinase: R Recombination site: RS 12-7-12 The recombination sites pair in same direction Inversion a b c d a b c d Deletion/ Integration Exchange/ Translocation Mutant sites LE/ RE mutation LE mutant RE mutant Spacer Mutation lox511 A ATAACTTCGTATA GCATACAT TATACGAAGTTAT TATTGAAGCATAT CGTATGTA ATATGCTTCAATA Reactions between mutant sites LE mutant X RE mutant LE+RE mutant + loxP loxP X lox511 lox511 X lox511 lox511 X lox512 Transposons Features. 1. Autonomous, may have a non-autonomous form. 2. Excise non-replicatively and re-insert elsewhere. Maize transposons: 1. Activator/Dissociation (Ac/Ds) 2. Enhancer/Suppressor-mutator (En/Spm) 3. Mutator (Mu) Maize Activator Element (Ac) TIR TIR TTT CATCCC TA AAAGTAGGGAT CAGGGATGAAA GT CCC TACTTT Exon 1 Exon 2 Exon 3 E4 E5 4565 bp mRNA ORF (2421 b) Outermost nucleotides of terminal inverted repeats are not complementary Ds element 1. Simple Ds elements are deletion derivatives of Ac, which have lost internal sequence for trans-acting factor. 2. Composite Ds elements internally contain rearranged Ac and unrelated sequences. Transposition 1. Insertion results in short target site duplication (“footprint”). This suggests that the mechanism of integration involves staggered cut (~8 bp) at the target site followed by strand synthesis. 2. Excision is imprecise and associated with nucleotide addition, deletion or inversion at their junction Requirements for transposition • • • Terminal inverted repeats (TIR) are essential for transposition. In addition to IR, ~240 bp sub-terminal nucleotides (both at 5’ and 3’ ends) of Ac are essential for transposition. At either ends of Ac internal deletions extending farther towards the termini results in gradual reduction in transposition. The element is immobilized when 116 bp or less at 5’ end and 102 or less at 3’ end are retained. Transposase (TPase) Characteristics of Transposition 1. Ac transposes by nonreplicative mechanism. It is physically excised from the donor site and re-inserted into the new position. 2. Ac transposes primarily, if not exclusively, during or shortly after replication. 2. Ac transposes from only one of the two daughter chromosomes. 3. Ac insertion may occur in both replicated or unreplicated target site. 4. Ac transposes preferentially to physically linked target site. a b Ac Ac Ac a Ac Ac b Ac Ac Ac 5’- and 3’- ends in direct orientation induce chromosome breakage One of the discoveries of McClintock was a phenomenon she called “breakage-fusion-bridge cycle”. Chromosome breakage occurred frequently at genetic locus, which she therefore called Dissociation (Ds). She later found that Ds locus could change its position in the genome, and that transposition of Ds and chromosome breakage required the presence of another locus, she designated Activator (Ac). Later other scientists elucidated that chromosome breakage is a consequence of aberrant transposition attempts between two different element ends in direct orientation. L R L R McClintock’s Ds locus Rarely transposes, often causes chromosomal breakage Strand selectivity model L R L R L After replication of two directly oriented left and right ends, one end on each daughter chromatid becomes incompetent. After DNA cleavage at the termini of the two competent ends, religation of the two ends leads to dicentric chromosome. Ac-TPase expression 1. 2. 3. Most of the Ac DNA sequence codes for for Ac-TPase. Ac promoter lacks a CAAT and TATA box and therefore is reminiscent mammalian housekeeping gene promoters. Like these, Ac promoter is weak and constitutively active. Ac-TPase structure • • • • • • It has 807 amino acids. The N-terminal contains 3 NLS. Between first and second NLS is TPase catalytic domain. Overlapping with NLS 2 and 3 is a DNA binding domain. TPase binds to sub-terminal, repetitive sequence motifs. DNA binding domain of TPase is methylation sensitive. Ac/ Ds can transpose in heterologous plant systems 1. tobacco. 2. Rice 3. Wheat 4. Arabidopsis 5. Tomato 6. petunia En/Spm transposable element system of maize Enhancer (En)/Suppressor-mutator (Spm) 13 bp perfect terminal inverted repeat P ATG ATG 8.3 kb tnpD tnpA Makes a 3-bp footprints (target site duplication) En/Spm Transposable elements cis Determinants for Excision: Approx. 180 bp at the 5’ end and 300 bp of 3’ end represent cis determinants. Contained in these regions are reiterations of a 12-bp sequence motif that is recognized by TNPA protein with 6 motifs present at 5’ end and 8 at 3’ end. Trans-factors: TNPA and TNPD (transposase complex) TNPA brings the two ends together and causes DNA bending, TNPD cleaves TNPA TNPD The “suppressor” function of En/Spm Transposable elements En/Spm have a unique feature: they act as suppressor of gene function The non-autonomous derivative of En/Spm (dSpm) when inserted into a gene causes reduced gene expression of that gene instead of knocking it out. The residual gene activity is due to the spicing of dSpm from pre-mRNA. However, if trans-factors TNPA is present then gene activity is knocked out i.e. pre-mRNA is not formed. TNPA binding with dSpm probably causes steric hindrance for RNA polymerase. Mutator Transposable element system of maize Mutator (Mu) trait was first identified by Robertson (1978) as a heritable high forward-mutation rate exhibited by maize lines. Many of these de novo mutations exhibited somatic instability, primarily apparent reversion to wildtype. This phenomenon was found to be associated with Mu transposable elements. Autonomous MuDR element (4942-bp): 200-bp TIR, create 9-bp target site duplication. TIR P 2.8-kb RNA (823 aa peptide) 1-kb RNA (207 aa peptide) Both proteins tightly bind to TIRs P TIR Several Mu elements (subfamilies) exist that contain variable internal sequence. These are non-autonomous derivatives of MuDR. Applications: 1. Mu elements are known to transpose to any locus, especially genes, therefore it is very useful for creating tagged mutations. 2. Mutator’s frequent transposition activity (even to unlinked locus) is reminiscent of P element system of Drosophila. In Drosophila, P elements have been used as vectors to increase the efficiency of transgene integration in the injected oocytes. Therefore, Mu is an attractive system for increasing transformation efficiency of maize. The strategy would involve introducing gene-of-interest in a Mu element and then injecting that into maize cells containing MuDR (MuDR based cloning in impossible because MuDR is unstable in E. coli). However, no report has so far been published that describes the success for this strategy.