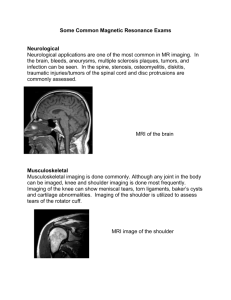
Nuclei most commonly used in in vivo NMR
advertisement

Supplemental File 1 - Basic information on MRI Nuclei most commonly used in in vivo NMR Especially due to their importance and relative high concentration, the nuclei shown in Table 1 are most commonly used in in vivo MRI. The Larmor frequencies at 17.6T of the listed nuclei are provided to illustrate the enormous frequency range. Table 1: NMR properties of nuclei most commonly used in in vivo NMR Isotope Spin γ/[MHz/T] Frequency @17.6 T /[MHz] natural abundance/ [%] 1H 2H 13C 19F 23Na 31P 39K 1/2 1 1/2 1/2 1 1/2 1/2 1 1/2 42.58 6.54 10.71 40.08 11.27 17.25 1.99 750 115 189 706 198 304 35 99.985 0.015 1.108 100 100 100 4.672 Standard elements of an MRI experiment In general a MRI experiment can be divided in three parts, slice selection, phaseencoding and frequency-encoding as shown in a standard two-dimensional gradient echo sequence (Fig. S1A). - slice selection: The combination of a frequency selective 90o RF pulse with a frequency bandwidth of z and a magnetic gradient selectively excites protons within a slice thickness given by z: z z Gslice The slice thickness can be derived by looking at the spatial dependency of the Larmor frequency when an additional linear magnetic field gradient in z direction is applied: ( z ) ( B0 zG z ) The opposite gradient following the slice selection shown in Fig. 1A is refocusing the signal. 1 - phase encoding: The phase encoding gradient G y imparts a spatially dependent phase shift into the signal. The whole experiment has to be repeated N PE times with different Gy values to cover the whole k space. ky - G y PE 2 frequency encoding: During data acquisition the frequency encoding gradient generates a spatially dependent precessional frequency in the acquired signal. kx Gxt 2 The opposite gradient shown in Figure 1A in the frequency encoding direction has the purpose to dephase the signal before it gets rephrased during the acquisition. In each repetition a whole k-space line with NFE points is acquired (see Fig. S1B). A Spin echo pulse sequence 900 1800 B k – space scheme ky echo RF slice Gz NPE freq. Gx NFE acqu. kx phase encoding (NPE) phase Gy TE frequency encoding (NFE) TR Figure S1. The left panel (A) shows the most commonly used T2 weighted spin echo (SE) sequence. The schematic representation of a two dimensional k-space is shown on the right (B). The light grey arrow indicates the acquisition of all frequency encoding points during one repetition (repetition time: TR) with the phase encoding gradient being at its third step in positive direction. 2 Standard imaging contrast To illustrate the different contrasts achievable with MRI a slice of a carrot tap root was acquired with different imaging parameters using standard imaging sequences. Example of relaxation times for protons in plants at 11.75 tesla are 1.5 s for T1, 15 ms for T2 and 5 – 10 ms for T2* respectively (Köckenberger, 2001). An image is referred to being T1 weighted if the TR is kept relatively short. While tissues with long T1 values will produce a hypo intense signal, tissues with shorter T1 will appear brighter. The overall signal intensity of the image will be reduced compared to no T1 weighting (TR = 5 x T1). Figure S2A shows the same slice of the carrot tap root acquired with different repetition times (TR = 10s, 2s, and 0.5s). The signal loss from A(i) to A(iii) is evident and is equal to a loss of signal to noise ratio (SNR) from 129 in A(i) to 36 in A(iii). To increase the visibility of A(iii), the brightness of the image was adjusted in A(iv). Compared to the non T1 weighted image A(i), the meristematic tissue in this image (white arrow) is much more pronounced, indicating shorter T1 values compared to the surrounding tissue. T2/T2* weighting is achieved by extending the echo time. With a longer TE the time for relaxation increases and tissues with shorter T2/T2* times will generate a more hypo intense signal compared to the ones with longer T2/T2*. The before mentioned GE sequence produces T2* weighting rather than T2 weighting in the images as the relaxation of M xy due to the inhomogeneous B 0 is not recovered. An application of an (slice selective) 180 degree RF pulse in the middle of TE has the ability to refocus those losses. This sequence (based on the Hahn spin echo) is therefore called spin echo sequence and is the most commonly used sequence for T2 weighting. T2 weighted images from a spin echo sequence are shown in Fig. S2B. With increasing echo time (TE = 9.5, 15, and 25 ms) the areas with shorter T2 are getting darker (arrows) compared to the surrounding tissue. Fig. S2C displays images from the same slice acquired with a GE sequence (TE = 2.6, 5, 10 ms). It can be seen that the signal is reducing much faster at much shorter TE compared to the SE images indicating T2* instead of T2 weighting. While the side root in the SE images at a TE of 25 ms is still visible (Fig. S2B(iii)), it can hardly bee identified at 10 ms TE in the GE image (Fig. S2C(iii)). The image closest to a proton density weighted image is Fig S2A(i) (same as S2B(i)) as the TR is very long and only a modest T2 weighting was applied. As TE 3 in standard pulse sequences cannot be reduced to zero and a TR of 5 x T1 results in enormous imaging times, most of the MRI images are both T1, as well as T2/T2* weighted, depending on the desired contrast, and therefore the imaging parameters used, one weighting might be chosen to be bigger than the other. Figure S2. Images of the same slice of a carrot tap root. While the first two rows are SE images the third row are GE images with different image weighting. A(i)A(iii) show the effect of T1 weighting (TE= 9.5ms; TR= 10s, 2s, 0.5s). A(iv) is the same image as A(iii) with an increased image intensity. B(i)-B(iii) illustrate different T2 (TR= 10s; TE=9.5, 15, and 25ms) and C(i)-C(iii) different T2* (TR=100ms; TE=2.6,5,10ms) weightings in the image. Besides the mentioned weightings additional signal reduction is present in every image due to diffusion effects and susceptibility differences within the sample (see: ‘Image resolution and its limits’). Furthermore, quantitative parameter maps showing the distribution of MRI parameters can be generated. The most common maps display the distribution of the equilibrium magnetization M0, the T1, the T2 and the T2* times. To generate such a map the data needs to be acquired in a certain way. A multi spin echo 4 sequence (RARE, Hennig et al., 1986) for example can be used to acquire a T2 map. Instead of using the acceleration factor of the sequence (see Supplement 2) each echo is used to generate its own image. As the phase encoding gradient is changed after each echo train (N echoes) and not after each echo, the resulting imaging time increases by a factor of N. The resulting N images can be fitted on a pixel by pixel basis to the following equation: S (t ) S0 exp( t / T2 ) While S0 is the signal at TE=0, t represents the different echo times of the N images. Solving the equation for T2 reveals the T2 values of the sample in each pixel. Example of such T2 maps on the developing barley grain can be found in Glidewell et al (2006). As discussed in the next paragraph additional signal reduction caused e.g. by diffusion effects and magnetic susceptibility changes within the sample need to be considered especially when moving towards high resolutions. Instead of T2 values often apperent T2 values are reported. A comprehensive overview how the field strength B0, the spatial resolution and the echo time influences the observed T2 values can be found in Edzes et al (1998). Image resolution and its limits In general, the voxel size of a two dimensional MRI image is defined by the slice thickness and the ‘in plane’ resolution x and y: x FOV x 1 1 2 Nx N x k x k x ,max G x Tacq y FOV y Ny 1 1 2 N y k yx k y ,max N y G y ,max PE While in this example Nx is the number of frequency and N y is the number of phase encoding points, FOV x and FOVy are the field of views in the corresponding directions. The maximum k x,max is reached with t = T acq (Tacq = acquisition time). It needs to be noted that MRI is not limited to two dimensional imaging, a real three dimensional dataset can be achieved by applying an additional phase encoding (NPE2) in the slice selection direction. If the resolution is not high enough to resolve the desired structure, or if the voxel lies on an edge of a compartment within the specimen, the voxel intensity is a mixture from signals of different tissues. This ‘partial volume’ effect will result in a 5 blurred image and real structures might not get resolved. An example is shown in Fig. S3. Although the images are not acquired at the exact same location it is obvious that structures resolved in the higher resolution image, are blurred in the low resolution one. Figure S3. High (left panel) and low (right panel) resolution images of a carrot tap root. Due to the partial volume effect the structures within the low resolution image appear blurred. Due to the T2 relaxation during the acquisition of the data (frequency encoding direction) the point spread function for each pixel will have a line width which also influences the resolution. If the point spread functions of two adjacent pixels does not fulfil the Rayleigh criterion the two pixels cannot be separated. As the T2 times in plants are usually relatively short and therefore yield large line widths (FWHM = 1/T2) the bandwidth (BW) of the experiment needs to be chosen to make sure that the T2 broadening is less than one pixel. As plants have many air pockets susceptibility effects in T2* weighted imaging can lead not only to a reduced resolution but also to image distortions. A good review about susceptibility effects on resolution is given by Callaghan et al. (1994). Another effect that needs to be taken into account is the diffusion of the spins out of the voxel in the frequency encoding direction during the acquisition of the signal Tacq. Although the effect is almost negligible at bigger voxel sizes, it needs to be 6 considered and can be substantial in smaller voxels. The average displacement <x> of, in our example, water molecules undergoing Brownian motion with a selfdiffusion constant DH20 = 2.23 x 10-9 m2s-1 at 20 oC can be written as: x 2 D Tacq To achieve a resolution x of 10 m with a FOVx of 4 mm e.g., N x = 400 points need to be acquired in the frequency encoding direction. Assuming a typical bandwidth (BW) of 50 kHz will result in an acquisition time of T acq = Nx/BW = 8 ms. With a resulting mean diffusion length of 6 m the average displacement <x> is already 60% of the voxel size resulting in substantial attenuation of the signal. According to the equation for resolution (x) calculations the necessary gradient strength Gx would be 1.8 T/m which exceeds already the capability of most commercially available gradient sets and shows another limit for higher resolution. To reduce the necessary gradient strength the T acq needs to be extended resulting in an even bigger signal loss due to diffusion. These remarks show that by increasing the resolution, besides the fact that less spins are within one voxel resulting in a lower SNR, new problems arise that need to be taken into consideration. SNR improvements Except moving to a lower resolution other measures can be applied to improve the SNR. Moving to a stronger magnet will in general increase the SNR by a substantial amount. While at lower field strength the theoretical increase is proportional to B 07/4 at high field strength the SNR gain tends to become closer to linear to B 0. As plants have a lot of susceptibility changes within their tissue mostly due to air enclosures using stronger magnets is not always advantages as additional artefacts can arise (T2*). Using an adapted RF resonator is another measure to increase the SNR. Besides using the before mentioned cryoprobes many other possibilities are available to construct an optimized RF resonator. Adapting the size of the RF resonator to ‘just fit’ the sample increases the SNR (high filling factor). Furthermore, smaller resonators have a higher sensitivity than larger ones and therefore achieve a higher SNR. While volume resonators like a solenoid or a birdcage resonator can be used for small samples (e.g. seeds, leaves, pods) a surface resonator like a simple single loop resonator is more appropriate for studies of small structures on larger plants 7 (e.g. the bark of a tree). Adding a preamplifier right next to the surface resonator increases the SNR furthermore. The need of a switch between transmission and reception of the signal of this setup would complicate the construction by a fair amount. The most straight forward way to increase the SNR is to use averaging and the appropriate (fast) imaging sequence. Chemical shift and principles of chemical shift imaging The electrons surrounding a nucleus produce a magnetic field which is in the opposite direction to B 0. This field, which is proportional to the static magnetic field B0, shields the nucleus from B 0 and the resonance frequency of the nucleus changes slightly depending on the strength of the additional field. Nuclei in different chemical environments (different positions in a molecule) have distinct but different resonance frequencies. The effective magnetic field at the nucleus is described by: B B0 (1 ) where is the shielding constant. In this way, different molecules can be identified in a NMR spectrum which is the result of a FID after a Fourier transformation. As the frequency shifts are usually very small is measured in parts per million (ppm). For in vivo proton NMR spectroscopy all the peaks that occur in a spectrum are in the range of about 12 ppm. By applying a water suppression technique (e.g. CHESS (Haase et al, 1985), or VAPOR (Griffey et al, 1990) the dominant water resonance can be suppressed and the resonances from much lower concentrated sugars or amino acids for example can be detected. In a standard three dimensional chemical shift imaging (CSI, Brown et al, 1982) experiment all three directions are phase encoded and a FID is acquired without any gradients present. The result is a four dimensional dataset in which each pixel of the three spatial dimensions contains a NMR spectrum. Integrating a peak allows to construct a three dimensional distribution of the observed resonance (metabolite map). The biggest trade-off of a CSI experiment is the long acquisition time since every point in k- space has to be acquired in a different transient and the relative low resolution due to the low metabolite concentrations. References 8 Brown T.R., Kincaid B.M., Ugurbil K. (1982) NMR chemical shift imaging in three dimensions Proc. Natl. Acad. Sci. USA 79, 3523-3526. Callaghan, P.T. (1994) Principles of Nuclear Magnetic Resonance Microscopy. Clarendon Press, Oxford, England. Edzes, H.T., van Dusschoten, D., and Van As, H. (1998) Quantitative T2 imaging of plant tissues by means of multi-echo MRI microscopy. Magn. Reson. Imaging 16, 185-196. Griffey, R.H., and Flamig, D.P. (1990) VAPOR for solvent – suppressed, short echo, volume – localized proton spectroscopy. J. Magn. Res. 88, 161-166. Haase, A., Frahm, J., Hanicke, W., and Matthaei, D. (1985) 1H NMR chemical shift selective (CHESS) imaging. Phys. Med. Biol. 30, 341-344. Köckenberger, W. (2001) Nuclear magnetic resonance micro-imaging in the investigation of plant cell metabolism. J. Exp. Bot. 52, 641-652. 9