Modern System of Bacterial Taxonomy
advertisement

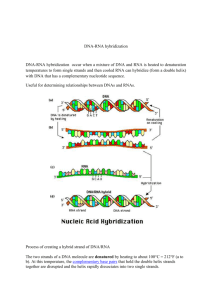
•DNA ANALYSIS •NUCLEIC ACID HYBRIDIZATION MODERN SYSTEMS OF BACTERIAL TAXONOMY Traditional vs Modern Binomial taxonomy DNA sequences Modern systems The routine laboratory identification are the microscopic observation, culturation biochemical test and serology test Latest and the most modern era of identification are molecular biological test Most of the methods relates to the content of genetic materials Serology Studies of serum and immune responses thru serum m/o is antigenic, normally response as antibodies The reagent used in test kit is antiserum Present of m/o usually causing the clotting effects towards antiserum (+ve result) Oppositely, the absent of m/o will be diluted effects (-ve result) Types of serology test Slide agglutination test ELISA (Enzyme-linked Immunosorbent Agglutination) Western blotting (same approach but not categorize as serological test) Phage typing To determine which phages a bacterium susceptible to Eg. Bacteriophage (the bacterial virus) only attacking the particular strain or types of bacteria Fatty acid analysis To compare the fatty acid profiles among m/o Also known as FAME (fatty acid methyl ester) Widely used in clinical and public health laboratories Flow cytometry To identify bacteria in sample without culturing the bacteria Cell and m/o go through a small portion Pass thru a laser and show scattering graph determining size, shape, density and surface Also consist of fluorescent detection DNA base composition Show the relatedness via % of (Guanine G and cytosine C) G complement with C, A complement with T Pair s of GC resulting pairs of AT; (GC + AT = 100%) Eg. If there are 2 m/o, might be 1st have 40%GC but the other have 80%GC DNA Fingerprinting To determine the source of hospital-acquired infections Spread via finger tips Relate to hospital personnel or patients from the same ward Short-time period required, instead of routine process Should have the control sequences Similar sequences shows >% of relationship Polymerase Chain Reaction m/o cannot be cultured through conventional methods This method can be used to increase the amount of microbial DNA to levels that can be tested by gel electrophoresis Eg. From the amber of ancient period If there are primer of particular m/o in the sample, there will be amplified DNA indicates that m/o, obviously shows in the gel 5 minutes break Nucleic acid hybridization •Southern Blotting •DNA chips •Ribotyping and Ribosomal RNA sequencing •Flourescent In Situ Hybridization (FISH) Introduction Each complimentary DNA containing complementary bases When heat, the hydrogen bonds separated When cool, it reunite almost back to normal Basic purpose is to identify unknown m/o Can hybridize any single-stranded nucleic acid chain; DNA-DNA, RNA – RNA, DNA-RNA Nucleic acid hybridization Nucleic acid hybridization Southern Blotting To identify unknown m/o Using DNA probe for rapid identification Method involves breaking enzyme, then selecting a specific fragment as a probe , then the probe able to hybridize with the DNA of all particular m/o strains, but not with the DNA of closely related group m/o DNA probe used to identify bacteria Southern Blotting Arrangement of papers in Blot tray. Southern blot DNA chips Possible to quickly detect a pathogen in a host by identifying a gene that is unique to that pathogen The chips composed of DNA probe Unknown DNA label by fluorescent dye and placed in a chips Hybridization between DNA probe and DNA detect by fluorescent DNA chip technology Fluorescent In Situ Hybridization Fluorescent dye-labeled RNA or DNA probe are used to specifically m/o in place or in situ Cell will be treated then the probe will enter the cell easily and react to the target ribosome in the cell (in situ) To determine the identity, abundance and relative activity of m/o in an environment Also to detect bacteria that have not yet been cultured (tiny quantity) FISH Ribotyping and Ribosomal RNA Sequencing To determine the phylogenic relationships among organisms 3 advantages: All cells contain ribosomes RNA genes undergo <changes over time – each sequences looks ‘same’ to each other Cell do not have to be cultured in laboratory Ribosomal sequencing Task of the week Compare between Western Blotting and Southern Blotting. Differentiate in table form. SCORE A!!! THE END