Perfinity Mass Spec Sample Prep
advertisement

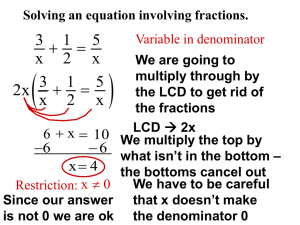
The Perfinity Workstation What about function and activity? Isoforms PTMs CD44 Non-small cell lung cancer A single insertion differentiates metastatic from benign cancer Kinase (enzyme) stuck in “on” position Unregulated growth TFSTVHPIPDE Prostate Specific Antigen (PSA) 61-78% false positive rate by immunoassay At least 4 isoforms Mikolajczyk et al. (2004), Clin. Biochem. 37:519-528. E su rfa Antibodies have trouble differentiating between little differences in proteins ce How serious is the isoform problem? There are ~30,000 genes in the human genome. Humans express >300,000 protein species. Proteins can exist as 10-50 isoforms simultaneously, many differing in biological activity. Single amino acid variants can cause a disease. Federal regulatory agencies are concerned about which isoforms are biologically active and whether biological averages of all isoforms are being reported. Differentiating between isoforms is an essential part of quantification and establishing structure-function relationships. Why Is There Not Broad Application? Protein sample preparation bottleneck Current approaches are a) Complex b) Multifaceted c) Often irreproducible Automating Sample Preparation Delivers… 1) Quality – Exceptional reproducibility!!! 2) Speed – 4 minute digests 3) Value – Costs ½ as much Automation of Established Technologies/Best Practices ClK K R Na+ Na+ R Cl- ClNa + The Workstation Platform 1. Two, independent centrally controlled pumping systems. 1 1 0 1 1 0 6 6 1 1 0 Quat 6 PDA PDA UFLC Pump A Pump B UFLC FRC CBM UFLC 2. The pumping systems are capable of flow rates ranging from 10 uL/min to 10 mL/min and an operating pressure of 10,000 psi. 3. Column I.D. can range from 300 um to 4.6 mm. 4. Detection is occurring at multiple points in the analysis. 5. Auto-sampler holds reagents and samples 6. Sample analysis is totally automated. 7. Multiple patented technologies integrated into a single platform. Digestion of Insulin Under Standard Conditions uV 5000000 4500000 4000000 GFFYPTK Undigested Protein 3500000 30 minutes 3000000 1 hour 3 hours 2500000 2000000 6 hours 1500000 12 hours 1000000 500000 18 hours 24 hours 0 4.0 4.5 5.0 Protein Product Sample: Column : Mobile Phase A: Mobile Phase B: Detection: 5.5 6.0 5ug insulin HALO 2.1x100mm RPC 2% ACN, 98% water, 0.1% Formic Acid 90% ACN, 10% water, 0.1% Formic Acid UV/VIS at 214nm 6.5 7.0 min Introduces >10% Variability Slept in Couldn’t sleep 9 With Perfinity Complete Digest in 8 Minutes or Less uV 2750000 Data1:Digest Insulin 50uL 4-185-03 400uLmin 60C May 17 Run 1.lcd Detector A:214nm Data2:Digest Insulin 50uL 4-185-03 200uLmin 60C May 17 Run 3.lcd Detector A:214nm Data3:Digest Insulin 50uL 4-185-03 100uLmin 60C May 17 Run 2.lcd Detector A:214nm 2500000 Data5:Digest Insulin 50uL 4-185-03 50uLmin 60C May 17 Run 3.lcd Detector A:214nm 2250000 Undigested Protein 2000000 1750000 GFFYTPK 1500000 1250000 1 minute 1000000 2 minutes 4 minutes 8 minutes 750000 500000 250000 0 19.0 20.0 21.0 22.0 23.0 24.0 25.0 26.0 min Protein Product Sample: Column 1: Column 2: Mobile Phase A: Mobile Phase B: Detection: 5ug insulin Perfinity Optimized Trypsin Column HALO 2.1x100mm RPC 2% ACN, 98% water, 0.1% Formic Acid 90% ACN, 10% water, 0.1% Formic Acid UV/VIS at 214nm What about something a little more complex? uV Data1:10uL R&A Tr 4-139-03 2pt1mm Halo 1 x20 300A GRD R8 Apr 1.lcd PDA Ch1:214nm,4nm(1.00) Data2:10uL R&A Tr 4-139-03 2pt1mm Halo 1 x20 300A GRD R7 Apr 1.lcd PDA Ch1:214nm,4nm(1.00) Data3:10uL R&A Tr 4-139-03 2pt1mm Halo 1 x20 300A GRD R6 Apr 1.lcd PDA Ch1:214nm,4nm(1.00) -200000 Data4:10uL R&A Tr 4-139-03 2pt1mm Halo 1 x20 300A GRD R5 Apr 1.lcd PDA Ch1:214nm,4nm(1.00) Data5:10uL R&A Tr 4-139-03 2pt1mm Halo 1 x20 300A GRD R4 Apr 1.lcd PDA Ch1:214nm,4nm(1.00) Data6:10uL R&A Tr 4-139-03 2pt1mm Halo 1 x20 300A GRD R3 Apr 1.lcd PDA Ch1:214nm,4nm(1.00) Time -300000 Run 1 Run 2 Run 3 Average StDev CV(%) 20.0 77551 72199 80057 76602.33 4014 5.24 20.4 123633 106875 121037 117181.7 9020 7.70 20.9 101131 108713 105673 105172.3 3816 3.63 21.1 164803 168049 167296 166716 1699 1.02 21.5 261128 236951 234215 244098 14812 6.07 22.2 290171 290735 288299 289735 1275 0.44 22.4 186600 178884 178861 181448.3 4461 2.46 22.7 143964 144298 142517 143593 947 0.66 22.9 104212 114875 110346 109811 5352 4.87 23.2 267686 255772 249757 257738.3 9125 3.54 23.4 130573 124909 129993 128491.7 3116 2.43 23.5 53220 48288 49150 50219.33 2634 5.25 23.6 31610 32296 31467 31791 443 1.39 23.8 102430 105401 101824 103218.3 1914 1.85 24.0 49022 56231 53091 52781.33 3614 6.85 24.2 91279 86549 78148 85325.33 6650 7.79 24.4 85161 85883 82149 84397.67 1981 2.35 24.8 241992 243983 250666 245547 4544 1.85 25.0 483586 471184 467367 474045.7 8480 1.79 25.5 34326 36274 31764 34121.33 2262 6.63 -400000 -500000 -600000 -700000 -800000 -900000 -1000000 20.0 22.5 25.0 27.5 30.0 32.5 35.0 37.5 40.0 •Reduced and alkylated transferrin •CV’s of first 20 peptides of first 3 runs Average CV = 3.69% 42% Sequence Coverage Sample process time = 44 minutes 42.5 min Average CV = 3.69 How does it compare? Time uV -880000 Data1:Solution Dig Transferrin 100uL 4-158-04.lcd PDA Ch1:214nm,4nm(1.00) Data2:Solution Dig Transferrin 100uL 4-158-06.lcd PDA Ch1:214nm,4nm(1.00) Data3:Solution Dig Transferrin 100uL 4-158-05.lcd PDA Ch1:214nm,4nm(1.00) -890000 -900000 -910000 -920000 Run 1 Run 2 Run 3 Average StDev CV(%) 16.55 852387 1149317 1161884 1054529 175173 16.61 17.00 3734784 4317305 4307224 4119771 333447 8.09 21.96 271377 298492 328634 299501 28642 9.56 22.99 428229 505473 520549 484750 49526 10.22 24.02 277636 311323 337541 308833 30030 9.72 24.35 451355 498854 561522 503910 55257 10.97 24.84 273835 316991 319944 303590 25811 8.50 25.73 304591 278655 314850 299365 18655 6.23 27.75 282650 346649 387876 339058 53022 15.64 29.39 276798 258535 270154 268496 9244 3.44 32.59 477172 557145 597765 544027 61357 11.28 33.55 288461 455468 509485 417805 115225 27.58 34.19 344019 271344 269006 294790 42650 14.47 34.58 510189 256581 319090 361953 132126 36.50 35.61 250712 291287 288096 276698 22561 8.15 36.17 386645 451144 489667 442485 52054 11.76 36.88 250506 280509 339414 290143 45230 15.59 37.41 762001 872341 920352 851565 81194 9.53 38.02 543185 680836 698035 640685 84875 13.25 39.35 580299 697772 742210 673427 83656 12.42 -930000 -940000 -950000 -960000 -970000 -980000 -990000 -1000000 -1010000 -1020000 15.0 20.0 25.0 30.0 35.0 40.0 45.0 min •Reduced and alkylated transferrin •CV’s of first 25 peptides of first 3 runs Average CV = 12.98% 20% Sequence Coverage Sample process time = 25 hours Average CV = 12.98 What about carry-over? uV -100000 Data1:10uL R&A Tr 4-139-03 1mm Halo 2pt1 x20 300A GRD no urea R1 Apr 1.lcd PDA Ch1:214nm,4nm(1.00) Data2:blank digestion run.lcd PDA Ch1:214nm,4nm(1.00) -125000 -150000 -175000 -200000 -225000 -250000 -275000 -300000 -325000 -350000 15.0 20.0 25.0 30.0 35.0 40.0 min How Did We Get Rid of Carry-over? By Maximizing Recovery!!! uV Data1:RAQ Hb S 25ug 4D 2-50 in 20 40C.lcd Detector A:214nm 500000 Data2:RAQ Hb S 25ug 4D 2-50 in 20 40C 50mM Tris.lcd Detector A:214nm 450000 400000 Perfinity Digest Buffer 350000 300000 250000 200000 150000 100000 50000 0 Standard Tris buffer -50000 -100000 -150000 10.0 12.5 15.0 17.5 20.0 22.5 25.0 min Confirmed uV Data1:RAQ Data2:RAQ 650000 Data3:RAQ Data4:RAQ 600000 Hb Hb Hb Hb S S S S 25ug 25ug 25ug 25ug 4D 4D 4D 4D 2-50 2-50 2-50 2-50 in in in in 20 20 20 20 40C.lcd Detector A:214nm 40C 50mM Tris.lcd Detector A:214nm 40C R 2.lcd Detector A:214nm 40C 50mM Tris R2.lcd Detector A:214nm 550000 500000 450000 400000 350000 300000 250000 200000 150000 100000 50000 0 7.5 10.0 12.5 15.0 17.5 20.0 22.5 min You are Taking An Uncompromising Approach High selectivity (+) •Antibodies can be used to isolate proteins from biological extracts. G G N N M M R L R L H H H H D D M A N Q M A N Q K K L L Poor Resolution (-) •Immunological contact areas (epitopes) are very small Chromatography Poor selectivity (-) •Proteins must be extracted or samples fractionated prior to analysis Reverse phase separation of insulin variants, some of which differ by a single amino acid. High resolution (+) •Similar proteins differing by small changes in structure can easily be resolved. High volume washing to minimize non-specific binding 1.6 120 Affinity sorbent stationary phase = -Protein G:Ab:Ag; 2 column volume wash. Antigen released from Ab with pH 2.5 eluent 1.4 100 1.2 Transferrin 0.8 60 Without abundant protein removal 0.6 80 Low affinity proteins are still bound to the protein G column. Immunoglobulins 40 0.4 20 0.2 0.0 0 0 2 4 6 8 10 12 14 16 Minutes 18 20 22 24 26 28 30 % AU 1.0 The value of high speed, high volume washing Affinity sorbent stationary phase = -Protein G:Ab:Ag; 40 column volume wash Antigen released from Ab with pH 2.5 eluent Transferrin The binding constant of transferrin is so much larger than the non-specifically bound proteins they are washed away at high wash volumes. Desorption of weakly bound proteins can occur with 60 to 120 seconds of washing. HSA A More Reliable Immunoassay A sample soln. [B-Ab:Ag] + -[Av4] Kd= 10-15M avidin H+ column -[Av4:B-Ab:Ag] = 1 fm avidin column -[Av4:B-Ab] + [Ag] RPC RPC trypsin digestion Universal affinity column Enhancing Capture Kinetics Antigen(s) eluting from avidin column. •Allow binding to occur in solution •Force antigens through an -Av tunnel transferrin RPC B-Ab:Ag B-Ab:Ag B-Ab:Ag B -Av Av-Av Av-Av Av- 100 nm - [Av] 100 – 1000 times larger than [B-Ab] minute Amount (ng) 5000 2500 1250 500 75 10 Standard Curve of Hu Transferrin Run 1 Run 2 Run 3 Average Stdev CV 210.1 225.8 222.8 219.6 8.3 3.8 202.6 211.0 218.8 210.8 8.1 3.8 175.9 178.1 179.5 177.8 1.8 1.0 131.6 130.5 130.9 131.0 0.6 0.4 62.7 62.6 64.4 63.2 1.0 1.6 28.7 30.2 27.9 28.9 1.2 4.0 Quantification using the Perfinity Workstation Sample Ag(1-10) R-Ab(1-10) RPC SILAC - Two samples are added to a single reaction vessel. Ab selection done at protein level P2 Label free/SRM. No change in standard work flow is needed. P3 P1 UV/Vis immunosorbent RPC B MALDIMS auto-sampler A UV/Vis ESI-MS waste oven nd 2 dimension LC refrigerated waste samples st 1 dimension LC MRM – Ab based selection can be done at either the protein or peptide level. Labeled peptides are added just before RPC. GIST/iTRAQ. Does not fit this work flow very well. Proteolysis and isotope coding must be done before Ab selection. This means all Ab selection is done at the peptide (or hapten) level. Quantitation Replicate Cal 10 Cal 100 Cal 1000 Cal 10000 1 486 1983 18688 209009 2 724 1970 19195 210932 3 637 1857 18595 204153 Mean: 615.6667 1936.667 18826 208031.3 StDev: 120.4256 69.29887 322.9288 3493.649 %CV: 19.60% 3.60% 1.70% 1.70% Assays with CV’s <10% uV Data1:R&A Data2:R&A Data3:R&A 1000000 Data4:R&A 1010000 Hb Hb Hb Hb A A A A 4-137-05 4-137-05 4-137-05 4-137-05 3 3 3 3 18 18 18 18 11 11 11 11 R4.lcd R3.lcd R2.lcd R1.lcd PDA PDA PDA PDA Ch1:214nm,4nm(1.00) Ch1:214nm,4nm(1.00) Ch1:214nm,4nm(1.00) Ch1:214nm,4nm(1.00) 990000 980000 970000 960000 950000 940000 930000 920000 910000 900000 890000 880000 870000 860000 17.5 Run 1 Run 2 Run 3 Run 4 Average StDev CV(%) 1 814 793 745 865 804 50 6.2 2 3 4 1355 942 1305 1283 933 1261 1269 874 1218 1428 1022 1284 1334 943 1267 73 61 37 5.5 6.5 2.9 20.0 5 1610 1336 1414 1601 1490 137 9.2 6 2469 2287 2310 2362 2357 81 3.4 22.5 Peak Number 7 8 9 2145 2353 1233 1803 2052 1131 1886 2084 1116 2064 2292 1223 1975 2195 1176 157 150 61 8.0 6.8 5.2 25.0 10 1535 1376 1485 1598 1499 94 6.3 11 1962 2005 1941 2245 2038 140 6.9 min 12 2805 2516 2648 2889 2715 166 6.1 13 714 649 664 789 704 63 9.0 14 439 378 393 454 416 36 8.7 15 1215 1012 1035 1156 1105 97 8.8 More of the Same Run 5 Run 10 Run 15 Run 20 Personalize Your Method Create your Batch 93% Sequence Coverage of Transferrin RT: 0.00 - 20.00 4.57 100 95 90 85 80 75 70 65 11.65 60 MRLAVGALLV FRDHMKSVIP LDAGLVYDAY KKDSGFQMNQ PRKPLEKAVA LNQYFGYSGA QYELLCLDNT LIWELLNQAQ FLKVPPRMDA VKWCALSHHE MNGEADAMSL PEAGYFAIAV IPMGLLYNKI RCLVEKGDVA LCLDGTRKPV RQQQHLFGSN HDRNTYEKYL CAVLGLCLAV SDGPSVACVK LAPNNLKPVV LRGKKSCHTG NFFSGSCAPC FKCLKDGAGD RKPVDEYKDC EHFGKDKSKE KMYLGYEYVT RLKCDEWSVN DGGFVYIAGK VKKSASDLTW NHCRFDEFFS FVKHQTVPQN EEYANCHLAR VTDCSGNFCL GEEYVKAVGN PDKTVRWCAV SEHEATKCQS KASYLDCIRA IAANEADAVT AEFYGSKEDP QTFYYAVAVV LGRSAGWNIP IGLLYCDLPE ADGTDFPQLC QLCPGCGCST VAFVKHSTIF ENLANKADRD HLAQVPSHTV VARSMGGKED NL: 1.42E8 FQLFSSPHGK DLLFKDSAHG Base Peak m/z= 300.00-2000.00 MS AIRNLREGTC PEAPTDECKP TransferrinOnlineDigest10uLof1 SVGKIECVSA ETTEDCIAKI 00ngmL-19Oct2011-1 CGLVPVLAEN YNKSDNCEDT DNLKGKKSCH TAVGRTAGWN GLNLCEPNNK EGYYGYTGAF TGGKNPDPWA KNLNEKDYEL APNHAVVTRK DKEACVHKIL FRSETKDLLF RDDTVCLAKL LRKCSTSSLL EACTFRRP 55 9.64 50 4.65 9.45 45 17.55 40 9.37 35 8.89 30 25 20 17.71 15 11.31 10 6.53 3.85 7.51 12.56 2.99 5 0.09 15.39 13.04 4.87 2.46 14.09 19.61 16.58 0 0 2 4 6 8 10 Time (min) 12 14 16 18 20 SRM of a specific peptide -MYLGYEYVTAIR RT: 0.00 - 20.00 RT: 0.00 - 20.00 10.86 100 4.57 100 NL: 1.42E8 Base Peak m/z= 300.00-2000.00 MS TransferrinOnlineDigest10uLof1 00ngmL-19Oct2011-1 95 95 90 90 85 NL: 1.88E6 m/z= 739.50-740.50 F: ITMS + c ESI Full ms [300.00-2000.00] MS TransferrinOnlineDigest10uLof100ngmL19Oct2011-1 MS2 Spectrum for 1 Peptide 85 80 80 75 70 75 65 70 60 55 11.65 65 9.64 50 4.65 60 45 9.45 17.55 40 55 9.37 35 8.89 50 30 25 45 20 17.71 40 15 11.31 10 6.53 3.85 2.99 35 5 0.09 0 2 12.56 30 4 14.09 15.39 13.04 4.87 2.46 0 7.51 6 8 10 Time (min) 12 19.61 16.58 14 16 18 20 25 20 15 9.66 14.73 10 6.27 5 2.84 4.57 5.45 3.02 9.48 9.94 8.95 7.11 11.57 11.70 14.28 12 14 15.62 16.82 18.22 0 0 2 4 6 8 10 Time (min) 16 18 20 Quantitation Standard Curve of Insulin C-terminal Peptide y = 20.843x - 537.99 R² = 0.9999 250000 200000 150000 100000 50000 0 0 2000 4000 6000 8000 10000 12000 Insulin Concentration (ng/mL) Replicate 10 100 1000 10000 Run 1 486 1983 18688 209009 Run 2 724 1970 19195 210932 Run 3 637 1857 18595 204153 Mean: 616 1937 18826 208031 StDev: 120 69 323 3494 %CV: 19.56 3.60% 1.70% 1.70% Isoform Identification MASCOT Results Rapid identification of VHLTPVEK sickle cell peptide isoform by IT-TOF •91% sequence coverage •Rapid identification of sickle cell variants •Fully automated extraction, digestion and analysis Selective Ion Monitoring A wide variety of oxidative stress induced posttranslational modifications (OSi~PTMs) can occur in the same protein at the same time. Carbonylation sites are revealed by MS/MS Y1 ion corresponds to the mass of biotinylated oxidized arginine (mass =374.19) Arginine oxidation product MS/MS Fragmentation of GLEWLGR*, a peptide from the immunoglobulin heavy chain variable region showing the oxidation of C-terminal arginine Distribution of the oxidized proteins in tissues of 30-35 year old males Number of proteins Most protein are in plasma as a result of cell death. In essence we are performing an autopsy on a cell that just died. Madian, A. G.; Regnier, F. E., Profiling Carbonylated Proteins in Human Plasma. J. Proteome Res. 2010, 9, (3), 1330-1343.