methods
advertisement

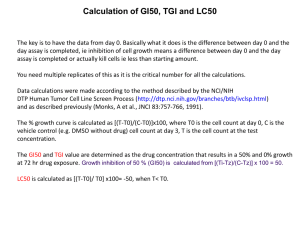
生物科技研究方法 Tools for analyzing gene expression Reference: fundamental molecular biology, Lizabeth Allison, 2007 蔡士彰 Central dogma DNA RNA protein Level Methods Transcription Northern blot In situ hybridization RNase protection assay Reverse transcriptase PCR (RT-PCR) Western blot ELISA Translation DNA-protein interaction EMSA, DNase I footprinting, Chromatin immunoprecipitation (ChIP) assay Protein-protein Pull down assay interaction Yeast two hybrid assay Coimmunoprecipition Fluorescence resonance energy trnsfer (FRET) RNA DETECTION & ANALYSIS **Steady-state levels of RNA** ** RNA levels that exist as a result of ** Transcription (+) versus degradation (-) METHODS: •Northern blotting •RNase protection assay •RT-PCR •Microarrays RNase Protection Assay +1 (start site) RPA +50 -100 T7 or SP6 pr omoter to make antisense RNA HindII Isite used to li near ize plasmid tr anscri ption bufferRNA , polymer ase , NTP's + [ -32P]UTP to make radiolabele d antisense RNA pr obe -100 +1 3' +50 5' hybr idize pr obe to purified mRNA Add RNase -100 3' +1 +50 5' 5' digest wi th RNaseT1 and RNase A (cleaves single-stranded RNAs) +1 AAAAAAA 3' mRN A +50 RNA duplex protected creates a RNA RNA hybr id den atur e an d analyze on a denatur ingpolyacr yl ami degel e G A T C >150 nt 50 nt RT-PCR mRNA Reverse transcription PCR Genome-wide comparison of mRNA expression between tumor and normal cells Microarrays Massively parallel analysis of gene expression • screen an entire genome at once • find not only individual genes that differ, but groups of genes that differ. • find relative expression level differences Effective for identify regulatory networks Reporter Assay Transfect embryo or ES cells DNA transfection methods 1. Calcium phosphate-DNA Coprecipitation method 2. DEAE-Dextran 3. Electroporation 4. Micro injection 5. Liposome Analysis of expression pattern in whole organism Direct enzymatic activity assay of cell lysate The major approaches of DNA delivery Physical delivery Chemical delivery Viral vectors Bactofection (bacterial vectors) Chemical delivery 1. Calcium-phosphate mediated transfection 2. polyplexes: eg. DEAE-dextran 3. Cationic liposomes: eg. Lipofectin, lipofectamine, lipofectase…. Physical delivery: electroporation, microinjection, gene gun (microballistics) Electroporation: Microinjection: Reporter genes X-gal IPTG: an inducer of β-galactosidase. This compound is used as a molecular mimic of allolactose Lactose is a disaccharide that consists of β-D-__________ and β-D-____________ molecules bonded through a β1-4 glycosidic linkage. X-gal is cleaved by β-galactosidase yielding galactose and 5-bromo4-chloro-3-hydroxyindole. 5-bromo-4-chloro-3-hydroxyindole then is oxidized into 5,5'-dibromo-4,4'-dichloro-indigo, an insoluble blue product. Luciferase is a generic name for enzymes commonly used in nature for bioluminescence. The name itself is derived from Lucifer, which means lightbearer. The most famous one is firefly luciferase from the firefly Photinus pyralis. In luminescent reactions, light is produced by the oxidation of a luciferin (a pigment), sometimes involving Adenosine triphosphate (ATP). The rates of this reaction between luciferin and oxygen are extremely slow until they are catalyzed by luciferase The reaction takes place in two steps: luciferin + ATP → luciferyl adenylate + PPi luciferyl adenylate + O2 → oxyluciferin + AMP + light The reaction is very energy efficient: nearly all of the energy input into the reaction is transformed into light. As a comparison, the incandescent light bulb loses about 90% of its energy to heat. luciferase (luc) systems firefly species Photinus pyralis Expressed luciferase catalyses oxidation of compounds called luciferans ( ATP-dependent process) These compounds emit fluorescense luminometer measurement Mice are injected with LUC+ salmonellas. Sensitive digital cameras allow non-invasive detection. For GT vectors pictures look the same Chloramphenicol acetyltransferase is a bacterial enzyme that detoxifies the antibiotic chloramphenicol. It is responsible for chloramphenicol resistance in bacteria. This enzyme covalently attaches an acetyl group from acetyl-CoA to the chloramphenicol molecule so that it is unable to bind to the ribosome. Reporter gene systems chloramphenicol acetyl transferase (CAT) CAT is a bacterial enzyme that catalyzes the transfer of acetyl groups from acetylcoenzyme A to the antibiotic chloramphenicol. (chloramphenicol deactivation) thin-layer chromatographic sheet Chloramphenicol is radiolabelled Green fluorescent protein (GFP) autofluorescent protein from Pacific Northwest jellyfish Aequorea victoria GFP is an extremely stable protein of 238 amino acids with unique post-translationally created and covalently-attached chromophore from oxidised residues 65-67, Ser-Tyr-Gly ultraviolet light causes GFP to autofluoresce In a bright green color Jellyfish do nothing with UV, The activate GFP by aequorin (Ca++ activated, biolumuniscent helper) GFP expression is harmless for cells and animals GFP construct could be used for construct tracking in living organism GFP labelled image of a human tumor. Vessel on the tumor surface are visible in black Level Methods Transcription Northern blot In situ hybridization RNase protection assay Reverse transcriptase PCR (RT-PCR) Western blot ELISA Translation DNA-protein interaction EMSA, DNase I footprinting, Chromatin immunoprecipitation (ChIP) assay Protein-protein Pull down assay interaction Yeast two hybrid assay Coimmunoprecipition Fluorescence resonance energy trnsfer (FRET) Level Methods Transcription Northern blot In situ hybridization RNase protection assay Reverse transcriptase PCR (RT-PCR) Western blot ELISA Translation DNA-protein interaction EMSA, DNase I footprinting, Chromatin immunoprecipitation (ChIP) assay Protein-protein Pull down assay interaction Yeast two hybrid assay Coimmunoprecipition Fluorescence resonance energy trnsfer (FRET) Electrophoretic-Mobility Shift Assay (EMSA) DNA binding site 20-30 nt probe + DNA binding protein Protein/DNA complex Electrophoretic-Mobility Shift Assay Incubate protein and DNA probe Load onto non-denaturing PAGE Resolve complexes & free probe protein concentration - shifted probe free probe + 0 EMSA Resolution of multiple protein/DNA complexes: Proteins sharing a DNA binding element G A G A 0 0 B Deoxyribonuclease I transcription factor 5' 3' 3' 5' * Deoxyribonuclease I 5' 3' 5' 3' 5' 3' 5' 3' 5' 3' 5' 3' 5' 3' 5' 3' 5' 3' 5' 3' 5' 3' 5' 3' 5' 5' 3' 3' G A G 0 0 DNAase I Footprinting Deoxyribonuclease I transcription factor 5' 3' 3' 5' Deoxyribonuclease I 5' 3' 5' 5' 5' 5' 3' 100-300 bp DNA fragment 3' with unique end-label3' 3' 5' 3' 5' 5' 5' 3' Incubate with test protein(s) 3' Digest3' with DNase I 5' 5' 3' Run3' digested DNA on PAGE 5' 3' 5' 5' 3' 3' Summary Methods to analyze transcription - detection of mRNAs - determination of transcriptional start sites -genome wide expression analysis by microarrays -reporter assay systems Methods to determine core promoter and/or enhancer elements -promoter mutagenesis -sequence alignment Methods to study DNA-protein interactions -EMSA, -DNase foot printing - Chromatin IP Level Methods Transcription Northern blot In situ hybridization RNase protection assay Reverse transcriptase PCR (RT-PCR) Western blot ELISA Translation DNA-protein interaction EMSA, DNase I footprinting, Chromatin immunoprecipitation (ChIP) assay Protein-protein Pull down assay interaction Yeast two hybrid assay Coimmunoprecipition Fluorescence resonance energy trnsfer (FRET)