EPIGENETIKA
SUMMER 2011
Petr Svoboda
mail:
tel:
svobodap@img.cas.cz
241063147
DNA METHYLATION I
(DNA METHYLATION AND ITS DETECTION)
EPIGENETICS
Epigenetics deals with heritable information
which is not encoded in the DNA sequence
Such information can be encoded in:
• structure and chromatin modifications
• DNA modifications
• RNA molecules
Bacteria - wide range of functions
Eukaryota - numerous effects
Protista, Plantae, (Mammalia?)
Bacteria - protection against RE
See Ratel 2006
Bacteria - protection against RE
Specific nucleotides are modified
-CcGG-GGcC-
all sites *
Hemophilus parainfluenzae
-cCGG-GGCc-
all sites *
Moricardia sp.
-GaTC-CTaG-
all sites (dam)
Escherichia coli
-Cc(A/T)GG-GG(T/A)cC-
all sites (dcm)
Escherichia coli
-cG-Gc-
-cNG-GNc-
some sites **
mammals, some fungi (Neurospora) and plants
some sites **
plants
* An example of a modified restriction enzyme recognition site. These sites are usually modified in organisms
with the corresponding restriction activity.
** Fraction of CG dinucleotides or CNG trinucleotides varies with species and, to a lesser extent, with tissue.
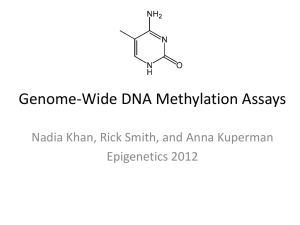
DNA methylation
5-methyl cytosine methylation
http://www.med.ufl.edu/biochem/keithr/research.html
Robertson 2002
Mammalian DNA methyltransferases
Maintenance DNA methylation
Ignore
HW: Why?
De novo DNA methylation
Methods to study DNA methylation
Global methylation analysis
HPLC (complete hydrolysis, AP)
TLC (complete hydrolysis, MspI, 32P labeling with PNK)
Sequence-specific methylation analysis
Methylation sensitive restriction enzymes - a number of methods
Bisulfite sequencing
MeDIP
For more details, see Oakeley, 1999
Bisulfite sequencing
HW:
Why isn’t 5mC
converted?
dsDNA is resistant to conversion!
From Oakeley, 1999
http://www.methods.info/Methods/DNA_methylation/Bisulphite_sequencing.html
Epitect Bisulfite Sequencing Kit
(Qiagen)
Classical protocol (bisulfite protocol.doc)
- starting material: cells or DNA up to 200 ng
- extremely sensitive (100 DNA molecules)
- based on agarose embedding
Drawbacks:
- time consuming (approx. 10-11 hours)
- low throughput (typically up to 8 samples/run)
- low yield (200 ng max/reaction)
Epitect
- starting material: DNA 1 ng - 2 mg
- sensititvity OK for most applications
- faster (cca 6 hours), throughput OK
Whatever …
The critical component are primers!!
http://www1.qiagen.com/Products/Epigenetics/Epitect/EpitectBisulfiteKit.aspx?ShowInfo=1
Model case:
L1 promoter methylation analysis
- active, autonomous, non-LTR class
- retrotransposition in cis
- the most abundant retrotransposon in
mammalian genomes
- 4-500 000 insertion in the human genome
- ~100 full length intact elements
- typically silenced in somatic cells
(hypermethylation)
5’ UTR ORF1
EN
ORF2
RT
AAAn
~6 kb
BISULFITE SEQUENCING STEP BY STEP
1) Find your sequence – NCBI Genbank and Pubmed
>L1 5’ UTR
GGGGGGAGGAGCCAAGATGGCCGAATAGGAACAGCTCCGGTCTACAGCTCCCAGCGTGA
GCGACGCAGAAGACGGTGATTTCTGCATTTCCATCTGAGGTACCGGGTTCATCTCACTA
GGGAGTGCCAGACAGTGGGCGCAGGCCAGTGTGTGTGCGCACCGTGCGCGAGCCGAAGC
AGGGCGAGGCATTGCCTCACCTGGGAAGCGCAAGGGGTCAGGGAGTTCCCTTTCCGAGT
CAAAGAAAGGGGTGACGGACGCACCTGGAAAATCGGGTCACTCCCACCCGAATATTGCG
CTTTTCAGACCGGCTTAAGAAACGGCGCACCACGAGACTATATCCCACACCTGGCTCGG
AGGGTCCTACGCCCACGGAATCTCGCTGATTGCTAGCACAGCAGTCTGAGATCAAACTG
CAAGGCGGCAACGAGGCTGGGGGAGGGGCGCCCGCCATTGCCCAGGCTTGCTTAGGTAA
ACAAAGCAGCAGGGAAGCTCGAACTGGGTGGAGCCCACCACAGCTCAAGGAGGCCTGCC
TGCCTCTGTAGGCTCCACCTCTGGGGGCAGGGCACAGACAAACAAAAAGACAGCAGTAA
CCTCTGCAGACTTAAGTGTCCCTGTCTGACAGCTTTGAAGAGAGCAGTGGTTCTCCCAG
CACGCAGCTGGAGATCTGAGAACGGGCAGACTGCCTCCTCAAGTGGGTCCCTGACCCCT
GACCCCCGAGCAGCCTAACTGGGAGGCACCCCCCAGCAGGGGCACACTGACACCTCACA
CGGCAGGGTATTCCAACAGACCTGCAGCTGAGGGTCCTGTCTGTTAGAAGGAAAACTAA
CAACCAGAAAGGACATCTACACCGAAAACCCATCTGTACATCACCATCATCAAAGACCA
AAAGTAGATAAAACCACAAAG
BISULFITE SEQUENCING STEP BY STEP
1) Find your sequence
>L1 5’ UTR
GGGGGGAGGAGCCAAGATGGCCGAATAGGAACAGCTCCGGTCTACAGCTCCCAGCGTGA
GCGACGCAGAAGACGGTGATTTCTGCATTTCCATCTGAGGTACCGGGTTCATCTCACTA
GGGAGTGCCAGACAGTGGGCGCAGGCCAGTGTGTGTGCGCACCGTGCGCGAGCCGAAGC
AGGGCGAGGCATTGCCTCACCTGGGAAGCGCAAGGGGTCAGGGAGTTCCCTTTCCGAGT
CAAAGAAAGGGGTGACGGACGCACCTGGAAAATCGGGTCACTCCCACCCGAATATTGCG
CTTTTCAGACCGGCTTAAGAAACGGCGCACCACGAGACTATATCCCACACCTGGCTCGG
AGGGTCCTACGCCCACGGAATCTCGCTGATTGCTAGCACAGCAGTCTGAGATCAAACTG
CAAGGCGGCAACGAGGCTGGGGGAGGGGCGCCCGCCATTGCCCAGGCTTGCTTAGGTAA
ACAAAGCAGCAGGGAAGCTCGAACTGGGTGGAGCCCACCACAGCTCAAGGAGGCCTGCC
TGCCTCTGTAGGCTCCACCTCTGGGGGCAGGGCACAGACAAACAAAAAGACAGCAGTAA
CCTCTGCAGACTTAAGTGTCCCTGTCTGACAGCTTTGAAGAGAGCAGTGGTTCTCCCAG
CACGCAGCTGGAGATCTGAGAACGGGCAGACTGCCTCCTCAAGTGGGTCCCTGACCCCT
GACCCCCGAGCAGCCTAACTGGGAGGCACCCCCCAGCAGGGGCACACTGACACCTCACA
CGGCAGGGTATTCCAACAGACCTGCAGCTGAGGGTCCTGTCTGTTAGAAGGAAAACTAA
CAACCAGAAAGGACATCTACACCGAAAACCCATCTGTACATCACCATCATCAAAGACCA
AAAGTAGATAAAACCACAAAG
HW:
look at CpG density of: EGFP, Bluescript, Actin promoter and transcribed region, IAP LTR, L1 ORF2
BISULFITE SEQUENCING STEP BY STEP
2) Word -> REPLACE CG WITH XY
>L1 5’ UTR
GGGGGGAGGAGCCAAGATGGCXYAATAGGAACAGCTCXYGTCTACAGCTCCCAGXYTGA
GXYAXYCAGAAGAXYGTGATTTCTGCATTTCCATCTGAGGTACXYGGTTCATCTCACTA
GGGAGTGCCAGACAGTGGGXYCAGGCCAGTGTGTGTGXYCACXYTGXYXYAGCXYAAGC
AGGGXYAGGCATTGCCTCACCTGGGAAGXYCAAGGGGTCAGGGAGTTCCCTTTCXYAGT
CAAAGAAAGGGGTGAXYGAXYCACCTGGAAAATXYGGTCACTCCCACCXYAATATTGXY
CTTTTCAGACXYGCTTAAGAAAXYGXYCACCAXYAGACTATATCCCACACCTGGCTXYG
AGGGTCCTAXYCCCAXYGAATCTXYCTGATTGCTAGCACAGCAGTCTGAGATCAAACTG
CAAGGXYGCAAXYAGGCTGGGGGAGGGGXYCCXYCCATTGCCCAGGCTTGCTTAGGTAA
ACAAAGCAGCAGGGAAGCTXYAACTGGGTGGAGCCCACCACAGCTCAAGGAGGCCTGCC
TGCCTCTGTAGGCTCCACCTCTGGGGGCAGGGCACAGACAAACAAAAAGACAGCAGTAA
CCTCTGCAGACTTAAGTGTCCCTGTCTGACAGCTTTGAAGAGAGCAGTGGTTCTCCCAG
CAXYCAGCTGGAGATCTGAGAAXYGGCAGACTGCCTCCTCAAGTGGGTCCCTGACCCCT
GACCCCXYAGCAGCCTAACTGGGAGGCACCCCCCAGCAGGGGCACACTGACACCTCACA
XYGCAGGGTATTCCAACAGACCTGCAGCTGAGGGTCCTGTCTGTTAGAAGGAAAACTAA
CAACCAGAAAGGACATCTACACXYAAAACCCATCTGTACATCACCATCATCAAAGACCA
AAAGTAGATAAAACCACAAAG
BISULFITE SEQUENCING STEP BY STEP
3) Word -> REPLACE C WITH T
>L1 5’ UTR
GGGGGGAGGAGTTAAGATGGTXYAATAGGAATAGTTTXYGTTTATAGTTTTTAGXYTGA
GXYAXYTAGAAGAXYGTGATTTTTGTATTTTTATTTGAGGTATXYGGTTTATTTTATTA
GGGAGTGTTAGATAGTGGGXYTAGGTTAGTGTGTGTGXYTATXYTGXYXYAGTXYAAGT
AGGGXYAGGTATTGTTTTATTTGGGAAGXYTAAGGGGTTAGGGAGTTTTTTTTTXYAGT
TAAAGAAAGGGGTGAXYGAXYTATTTGGAAAATXYGGTTATTTTTATTXYAATATTGXY
TTTTTTAGATXYGTTTAAGAAAXYGXYTATTAXYAGATTATATTTTATATTTGGTTXYG
AGGGTTTTAXYTTTAXYGAATTTXYTTGATTGTTAGTATAGTAGTTTGAGATTAAATTG
TAAGGXYGTAAXYAGGTTGGGGGAGGGGXYTTXYTTATTGTTTAGGTTTGTTTAGGTAA
ATAAAGTAGTAGGGAAGTTXYAATTGGGTGGAGTTTATTATAGTTTAAGGAGGTTTGTT
TGTTTTTGTAGGTTTTATTTTTGGGGGTAGGGTATAGATAAATAAAAAGATAGTAGTAA
TTTTTGTAGATTTAAGTGTTTTTGTTTGATAGTTTTGAAGAGAGTAGTGGTTTTTTTAG
TAXYTAGTTGGAGATTTGAGAAXYGGTAGATTGTTTTTTTAAGTGGGTTTTTGATTTTT
GATTTTXYAGTAGTTTAATTGGGAGGTATTTTTTAGTAGGGGTATATTGATATTTTATA
XYGTAGGGTATTTTAATAGATTTGTAGTTGAGGGTTTTGTTTGTTAGAAGGAAAATTAA
TAATTAGAAAGGATATTTATATXYAAAATTTATTTGTATATTATTATTATTAAAGATTA
AAAGTAGATAAAATTATAAAG
BISULFITE SEQUENCING STEP BY STEP
4) Word -> REPLACE XY WITH CG
>L1 5’ UTR
GGGGGGAGGAGTTAAGATGGTCGAATAGGAATAGTTTCGGTTTATAGTTTTTAGCGTGA
GCGACGTAGAAGACGGTGATTTTTGTATTTTTATTTGAGGTATCGGGTTTATTTTATTA
GGGAGTGTTAGATAGTGGGCGTAGGTTAGTGTGTGTGCGTATCGTGCGCGAGTCGAAGT
AGGGCGAGGTATTGTTTTATTTGGGAAGCGTAAGGGGTTAGGGAGTTTTTTTTTCGAGT
TAAAGAAAGGGGTGACGGACGTATTTGGAAAATCGGGTTATTTTTATTCGAATATTGCG
TTTTTTAGATCGGTTTAAGAAACGGCGTATTACGAGATTATATTTTATATTTGGTTCGG
AGGGTTTTACGTTTACGGAATTTCGTTGATTGTTAGTATAGTAGTTTGAGATTAAATTG
TAAGGCGGTAACGAGGTTGGGGGAGGGGCGTTCGTTATTGTTTAGGTTTGTTTAGGTAA
ATAAAGTAGTAGGGAAGTTCGAATTGGGTGGAGTTTATTATAGTTTAAGGAGGTTTGTT
TGTTTTTGTAGGTTTTATTTTTGGGGGTAGGGTATAGATAAATAAAAAGATAGTAGTAA
TTTTTGTAGATTTAAGTGTTTTTGTTTGATAGTTTTGAAGAGAGTAGTGGTTTTTTTAG
TACGTAGTTGGAGATTTGAGAACGGGTAGATTGTTTTTTTAAGTGGGTTTTTGATTTTT
GATTTTCGAGTAGTTTAATTGGGAGGTATTTTTTAGTAGGGGTATATTGATATTTTATA
CGGTAGGGTATTTTAATAGATTTGTAGTTGAGGGTTTTGTTTGTTAGAAGGAAAATTAA
TAATTAGAAAGGATATTTATATCGAAAATTTATTTGTATATTATTATTATTAAAGATTA
AAAGTAGATAAAATTATAAAG
BISULFITE SEQUENCING STEP BY STEP
5) Design primers
- size typically up to 500 bp, ideally around 300 bp
- avoid low complexity sequences
- Tm around 55oC
- select the right region!
>L1 5’ UTR
GGGGGGAGGAGTTAAGATGGTCGAATAGGAATAGTTTCGGTTTATAGTTTTTAGCGTGA
GCGACGTAGAAGACGGTGATTTTTGTATTTTTATTTGAGGTATCGGGTTTATTTTATTA
GGGAGTGTTAGATAGTGGGCGTAGGTTAGTGTGTGTGCGTATCGTGCGCGAGTCGAAGT
AGGGCGAGGTATTGTTTTATTTGGGAAGCGTAAGGGGTTAGGGAGTTTTTTTTTCGAGT
TAAAGAAAGGGGTGACGGACGTATTTGGAAAATCGGGTTATTTTTATTCGAATATTGCG
TTTTTTAGATCGGTTTAAGAAACGGCGTATTACGAGATTATATTTTATATTTGGTTCGG
AGGGTTTTACGTTTACGGAATTTCGTTGATTGTTAGTATAGTAGTTTGAGATTAAATTG
TAAGGCGGTAACGAGGTTGGGGGAGGGGCGTTCGTTATTGTTTAGGTTTGTTTAGGTAA
ATAAAGTAGTAGGGAAGTTCGAATTGGGTGGAGTTTATTATAGTTTAAGGAGGTTTGTT
TGTTTTTGTAGGTTTTATTTTTGGGGGTAGGGTATAGATAAATAAAAAGATAGTAGTAA
TTTTTGTAGATTTAAGTGTTTTTGTTTGATAGTTTTGAAGAGAGTAGTGGTTTTTTTAG
TACGTAGTTGGAGATTTGAGAACGGGTAGATTGTTTTTTTAAGTGGGTTTTTGATTTTT
GATTTTCGAGTAGTTTAATTGGGAGGTATTTTTTAGTAGGGGTATATTGATATTTTATA
CGGTAGGGTATTTTAATAGATTTGTAGTTGAGGGTTTTGTTTGTTAGAAGGAAAATTAA
TAATTAGAAAGGATATTTATATCGAAAATTTATTTGTATATTATTATTATTAAAGATTA
AAAGTAGATAAAATTATAAAG
BISULFITE SEQUENCING STEP BY STEP
5) Design primers
- size typically up to 500 bp, ideally around 300 bp
- avoid low complexity sequences
- Tm around 55oC
- select the right region!
>L1 5’ UTR
GGGGGGAGGAGTTAAGATGGTCGAATAGGAATAGTTTCGGTTTATAGTTTTTAGCGTGA
GCGACGTAGAAGACGGTGATTTTTGTATTTTTATTTGAGGTATCGGGTTTATTTTATTA
GGGAGTGTTAGATAGTGGGCGTAGGTTAGTGTGTGTGCGTATCGTGCGCGAGTCGAAGT
AGGGCGAGGTATTGTTTTATTTGGGAAGCGTAAGGGGTTAGGGAGTTTTTTTTTCGAGT
TAAAGAAAGGGGTGACGGACGTATTTGGAAAATCGGGTTATTTTTATTCGAATATTGCG
TTTTTTAGATCGGTTTAAGAAACGGCGTATTACGAGATTATATTTTATATTTGGTTCGG
AGGGTTTTACGTTTACGGAATTTCGTTGATTGTTAGTATAGTAGTTTGAGATTAAATTG
TAAGGCGGTAACGAGGTTGGGGGAGGGGCGTTCGTTATTGTTTAGGTTTGTTTAGGTAA
ATAAAGTAGTAGGGAAGTTCGAATTGGGTGGAGTTTATTATAGTTTAAGGAGGTTTGTT
TGTTTTTGTAGGTTTTATTTTTGGGGGTAGGGTATAGATAAATAAAAAGATAGTAGTAA
TTTTTGTAGATTTAAGTGTTTTTGTTTGATAGTTTTGAAGAGAGTAGTGGTTTTTTTAG
TACGTAGTTGGAGATTTGAGAACGGGTAGATTGTTTTTTTAAGTGGGTTTTTGATTTTT
GATTTTCGAGTAGTTTAATTGGGAGGTATTTTTTAGTAGGGGTATATTGATATTTTATA
CGGTAGGGTATTTTAATAGATTTGTAGTTGAGGGTTTTGTTTGTTAGAAGGAAAATTAA
TAATTAGAAAGGATATTTATATCGAAAATTTATTTGTATATTATTATTATTAAAGATTA
AAAGTAGATAAAATTATAAAG
3’ End dG should be above -13.0
(calculated from the last 7 nucleotides)
TTTT doesn’t matter
Duplex analysis
- ignore all dimers with positive dG
- only primers with negative dG <-1.5 should concern you
- combination of low negative dG and perfect basepairing at
the 3’ end is the worst combination
Tips how to improve amplification
HOT START PCR
-add Taq pol only after denaturation step
- use hot start Taq pol, e.g. Amplitaq GOLD from Perkin Elmer/ABI
- Amplitaq GOLD requires the intital denaturation step for 10-15 min at 95 oC
TOUCHDOWN PCR
94 oC for 15 min
94 oC for 30 sec
62->55 oC for 30 sec
72 oC for 1 min
14 cycles
0.5 oC down/cycle
94 oC for 30 sec
55 oC for 30 sec
72 oC for 1 min
36 cycles
72 oC for 15-20 min
EXTREMELY
SENSITIVE TO
CONTAMINATIONS
!!!
BISULFITE SEQUENCING STEP BY STEP
Restriction digest
- cheap and fast
- less information (up to a few CpGs)
- not good for polymorphic sequences
5) Run PCR
6) cut
7) Gel Extraction
(Qiagen)
8) TOPO TA II cloning
(Invitrogen)
9) Miniprep
(Qiagen)
Pyrosequencing http://www.pyrosequencing.com/
- short read (30nt)
- quantitative ratio of polymorphic nucleotides
- good for one sequence analyzed from many
samples
10) Sequencing with SP6 primer
Direction of sequencing is important!
-GT-rich regions are more difficult to sequence
- cause problems especially with products >300 bp
700
The same PCR product (amount, purity …) but different strands sequenced
400
BioEdit –it’s good and it’s free!
http://www.mbio.ncsu.edu/BioEdit/bioedit.html
Vector NTI is good but f*cking expensive
52
58 61
70
analysis of
individual
elements
single-locus
analysis
5’ UTR ORF1
AAAn
L1 Xq22
L1 Xp22
L1 6q16
L1 6p22
L1 6p21
L1 pool
EN
L1 8q24
ORF2
RT
female
undifferentiated
hES sample
?
?
?
How would you explain it?
How would you test your explanation?
5’ UTR ORF1
EN
8q24
Xp22
ORF2
RT
AAAn
HOMEWORK
Oct-4 promoter analysis
-PCR cloned in pCR II
-sequenced with SP6
MeDIP
DNA METHYLATION II
(EFFECTS OF DNA METHYLATION)
DNA METHYLATION ACROSS PHYLA
- a number of conserved genes
- common basic cell types
- highly variable DNA methylation
- different development
- conserved histone modifications
- different sex determination
- different epigenetic mechanisms
>350 MYA
>400 MYA
MAMMALIA
Mus
AMPHIBIA
Xenopus
PISCES
Danio
CHORDATA
>600 MYA
DEUTEROSTOMES
ECHINODERMATA
COELOMATES
PROTOSTOMES
Strongylocentrotus
ARTHROPODA
Drosophila
NEMATODA
Caenorhabditis
EUMETAZOA
PSEUDOCOELOMATES
EXTENSIVE DNA METHYLATION LEAVES TRACES ….
-genomes carrying CpG methylation show lower frequencies of CpG
Jabbari 2004
C-T CONVERSION
http://www.chemsoc.org/chembytes/ezine/2001/pufulete_mar01.htm
Drosophila
0.1-0.2% of the cytosines methylated
DNA methylation in the zebrafish
DNA METHYLATION DISTRIBUTION
- trends and exceptions
repetitive sequences
- noncoding tandem repeats (satellites)
- coding tandem repeats - rDNA
- interspersed elements - L1, IAP, Alu
- telomeric repeats
hypermethylated
variable
hypermethylated
hypermethylated
unique sequences
- promoter
- “gene body”
- active genes
- inactive genes
+/- unmethylated
variable
methylated
upstream of the UCE
H4Ac
(active)
UCE
core p.
rDNA
- pol I transcription
- tandemly arrayed on five pairs of
human acrocentric chromosomes
- ~400 copies per haploid genome
- typically half active, half inactive
(all active in the oocyte)
Methylation
hypo
hyper
dim-H3K9
(inactive)
HEK-293
total DNA
transformed cell lines
primary human cells
primary mouse cells
undiff. ES cells
diff. ES cells
50%
100%
100%
100%
100%
50%
0%
0%
0%
0%
400 bp
human
mouse
130 bp repeats
UCE Core
Annotation of UCE and CORE sequences based on Heix and Grummt , 1995
- different CpG density between closely related species
- gene activity does not correlate with methylation in both species
- only inactive genes in transformed lines acquire methylation
- sometimes, methylation is not very informative
CpG islands
Found in and near
approximately 40% of
promoters of mammalian
genes (about 70% in human
promoters). A “typical” CpG
island is 300-3000 bp long.
The CpG sites in the CpG
islands of promoters are
typically unmethylated if
genes are expressed. This
observation led to the
speculation that methylation of
CpG sites in the promoter of a
gene may inhibit the
expression of a gene.
Kim et al. BMC Cancer 2006 6:180
CpG island is a region least
200 bp long and with a GC
percentage that is greater than
50% and with CpG frequency
that is greater than 6%
(genome average is 1%).
PROMOTERS OF INACTIVE GENES
- hundreds of papers with contradictory data
- methylation correlates with inactivity … but that’s it ...
CpG poor promoters
-hypermethylated regardless of activity
strong CpG island promoters
-hypomethylated regardless of activity
weak CpG island promoters
-distinct …
testis-specific promoters
- methylated in somatic cells
Weber 2007
Weber 2007
Mammalian DNA methyltransferases
Maintenance methylation - DNMT1
substrate:
function:
hemimethylated DNA
restoration of DNA methylation after replication
De novo methylation - DNMT3a, 3b (3l)
substrate:
function:
unmethylated DNA
establishment of new DNA methylation patterns
Li 2002
Robertson 2002
Mammalian DNA methyltransferases
Maintenance DNA methylation
Ignore
HW: Why?
De novo DNA methylation
Cooperativity between
maintenance and de novo DNA methylation
LINE-1
IAP, centromeres
Cooperativity between
maintenance and de novo DNA methylation
Klose 2006
Setting up and interpreting the mark …
Klose 2006
Setting up and interpreting the mark …
DNA Methyl Binding Proteins MBDs
http://homepages.ed.ac.uk/dmac/Bird_Lab/birdlab.html
http://www.wcb.ed.ac.uk/bird.htm
MBDs
Klose 2006
Hendrich 2003
http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=OMIM
CHARACTERIZATION
MECP2
MBD1
MBD2
KO
MUTATIONS
-transcriptional repressor
- X-linked
-able to bind a single methyl-CpG
-binds tightly to chromosomes, pericentromeric heterochromatin
- associates with Sin3A HDAC complex and BRM complex
- expression correlates with maturation of the neural system
-mutant ES cells grow and can
differentiate (Tate et al., 1996)
-essential for embryonic
development (Tate et al., 1996)
-not essential for embryonic
development (Chen et al. 2001)
Rett syndrome
- various mutations
- only in females (lethal in m.)
-transcriptional repressor from the MeCP1 complex
-requires more than a single methyl-CpG
-in a complex with HDAC3
-interacts with the Suv39h1-HP1 heterochromatic complex
Mbd-/- mice have deficits in
adult neurogenesis and
hippocampal function (Zhao et
al., 2003)
Autism?
-transcriptional repressor from the MeCP1 complex
- also described as demethylase - but nobody reproduced that!
-MBD2/ NuRD complex, HDACs
-MBD2 recruiting the NURD complex to methylated DNA?
MBD4
Kaiso
Mbd2-/- mice are viable and
fertile! Maternal behavior of
Mbd2-/- mice is defective
(Hendrich et al. 2001)
-does not bind methylated DNA
-MBD3/ NuRD complex (distinct from MBD2 NURD)
-MBD3 an integral part of the NURD complex?
-Mbd3 -/- ES cells are viable
but fail to form a stable NuRD
complex, differentiation defect,
LIF-independent self-renewal.
- Mbd3-/- mice die during early
embryogenesis
-glycosylase domain, DNA repair
- interacts with mismatch repair protein MLH
-Mbd4 -/- mice have a 3-fold
increase in the frequency of Cto-T transitions at CpG sites
(Millar et al. 2002)
-lacks MBD domain
-recognizes 5mC in CGCG through zinc-finger domains
- transcriptional repressor
Kaiso -/-t mice show
resistance to intestinal cancer
MBD3
Angelmann syndrome?
Autism?
primary microsatelliteinstability (MSI) tumors
Klose 2006
Klose 2006
A few final notes regarding DNA methylation
- when it is associated with transcriptional repression, there are
usually also chromatin modifications consistent with the pattern
- information about DNA sequence methylation without information
about location (promoter, intergenic, intronic, exonic …) and
expression has low information content
- methylated promoter does not automatically belong to an inactive
gene while unmethylated promoter does not automatically belong to
an active gene
- it’s OK to say that DNA methylation is typically associated with
transcriptional repression
- be open minded and expect the unexpected ...
DNA DEMETHYLATION
- DNA methylation is stable but reversible
PASSIVE
- inhibition of DNMT1, - replication dependent
ACTIVE
- replication independent
- clearly demonstrated in some cases
- unknown mechanism, unknown demethylase. DNA repair/glycosylase?
CpG methylation during development
1-cell 6h
1-cell 8h
2-cell 22h
4-cell 45h
Adapted from Mayer et al. (2000). Nature 403(6769):501-2
4-cell asymmetrical staining
8-cell weak labeling, rare asymmetrical chr.
Adapted from Rougier et al. (1998). Genes Dev 12(14):2108 -13
NOBODY EVER REPRODUCED THESE DATA
THIS PAPER WAS NEVER RETRACTED
Reading the original literature ...
Introduction and Background:
What hypotheses are being tested in this paper?
What information made the authors to perform the experiments?
What new methods or insights brought to bear on the problem?
Methods:
What are the critical methods of the paper?
What enabling technologies are used?
What are the weaknesses of the methods used?
Are there other or better approaches that could be used?
If this is a genetics approach, what would be a biochemical or molecular approach?
If this is a biochemical or molecular approach, what genetics methods could be used?
Results and Discussion
What are the primary conclusions of the paper?
Did the authors test their hypotheses properly?
What novel information or directions come from this work?
What control experiments were performed?
What assumptions still remain in the work?
How could these assumptions be tested?
What other explanations for the observations are still possible?
HW: What’s the closest
mammalian homologue of
ROS1?