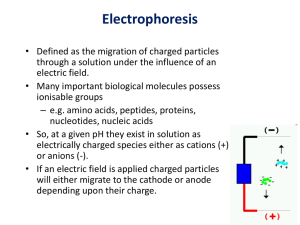
PowerPoint Presentation from May
advertisement
Protein Analysis F&S Core Pat Limbach, PhD CEG Protein Analysis F&S Core Objectives To enhance research in environmental proteomics by supporting proteomic services and technologies and: • Providing consultation and assistance with experimental design • Providing consultation on data interpretation by taking advantage of expertise in the Bioinformatics Core • Interfacing with the UC/GRI Proteomics and MS Facilities • Developing and disseminating capabilities (Protein Production, DIGE, Protein ID, PTM Identification, Biomarker Discovery) CEG Protein Analysis F&S Core Services Stimulate research of protein-environment interactions by providing a 50% subsidy for CEG members and Pilot Project awardees on: • 2-D Gel services (IEF, SDS, Staining, Imaging & Spot Picking) • Protein ID services (MALDI PMF, LC-MS/MS) • Protein characterization services (PTM, Biomarkers) • Bioinformatics (Modeling, Data interpretation) CEG Protein Analysis F&S Core CoM 2-D Gel Services/Capabilities Equipment/Capacity Genomic Solutions Investigator 2-D Electrophoresis System Genomic Solutions Gel Casting System Genomic Solutions ProImage Image Acquisition System Capacity: 40 IEF strips/wk; 20 SDS-PAGE gels/wk Services Complete 2-D Gel Service (with Spot Picking for MS Analysis) IEF Only SDS-PAGE (large format) Only Gel Staining (fluorescent and silver staining) SDS-PAGE Plus (includes staining & imaging) Image Analysis only or with spot picking CEG Protein Analysis F&S Core GRI 2-D Gel Services/Capabilities Equipment/Capacity 4 Amersham Multiphor II IEF units for 2-D gel electrophoresis 4 Genomic Solutions Investigator units for large scale 2-D gels 5 Invitrogen XCell units for small scale 1-D and 2-D gels Amersham ImageScanner imaging densitometer for gel scanning Compugen Z3 software for 2-D gel analysis Amersham Åkta Purifier10 HPLC system for non-gel protein analysis Services 1-D gel electrophoresis (small scale to medium scale) 2-D gel electrophoresis (small scale (~18 gels/wk) and large scale (~40 gels/wk)) Silver staining and Coomassie Blue staining Imaging densitometry of protein gels & comparative data analysis of 2-D gels Western blotting CEG Protein Analysis F&S Core GRI Mass Spec Services/Capabilities Equipment Finnigan LCQ Deca XP Max with Dionex Ultimate nanoflow 2-D HPLC Bruker Biflex III MALDI-TOFMS Sciex API 3000 Triple quadrupole ESI-MS with Eldex MicroPro microflow HPLC VG Fisons Quattro Single quadrupole ESI-MS Services MALDI Protein ID by Peptide Mass Fingerprinting LC-MS/MS Protein ID by Sequence Tag 2-D LC-MS/MS Protein ID of Mixtures Peptide/Metabolite Quantification Protein/Peptide/Oligonucleotide Purity Confirmation CEG Protein Analysis F&S Core UC Mass Spec Services/Capabilities Equipment/Capacity Waters Q-TOF II LC-MS with Waters Capillary HPLC Bruker Reflex IV MALDI-TOFMS Thermo LTQ-FTMS Services MALDI Protein ID by Peptide Mass Fingerprinting LC-MS/MS Protein ID by Sequence Tag Accurate Mass Measurements of Proteins Phosphorylation Analysis by IMAC (Magnetic Bead) Purification & MALDI and/or LC-MS/MS analysis (Protocol developed through CEG funding) Biomarker Discovery by ClinProt Magnetic Bead Purification & MALDI-MS (Protocol developed through CEG funding) CEG Protein Analysis F&S Core GRI Protein Production Services/Capabilities Equipment Biostat C-DCU CellTrol Äkta Explorer Services Subcloning of gene of interest into expression construct and sequencing Optimization of fermentation growth conditions Fermentation of the bacterial (12 L) or insect cell (4-32 L) culture containing the gene of interest Preparation of cell-free extracts Optimization of purification method, purification of the protein of interest (Quantities typically range from 10 mg to over 1 g depending on conditions) Personnel DJ Ferguson, PhD (GRI) CEG Protein Analysis F&S Core Overview of Process CEG Investigator Protein Analysis F&S Core Leader Bioinformatics Consultant Sample Submission Facility Cores (Protein Production Facility) (2-D Gel) (MS Services) Data/Product Generation CEG Investigator Facility Personnel Bioinformatics Consultant CEG Protein Analysis F&S Core Step 1: Information Required The following information will be required to insure the researcher obtains appropriate F&S Core services: 1. Sample type? purified protein, simple mixture (less than 10 proteins), or complex mixture (e.g., cell lysates, biological fluid like serum) 2. Amount of sample? 3. Information desired? relative protein abundances or comparative analysis with no identification of proteins, relative protein abundances or comparative analysis with subsequent identification of proteins, protein identification only, or protein identification plus additional characterization of protein modification(s) CEG Protein Analysis F&S Core Step 2: Initial Consultation Once initial information is obtained: • Contact Protein Analysis F&S Core Leader to schedule consultation • During consultation, appropriate facilities and services will be determined • Investigator will be instructed on sample submission and appropriateness and mechanisms of CEG supplement CEG Protein Analysis F&S Core Step 3: Data Review After services have been obtained • Facility staff will work with investigator on obtaining results • When necessary, consultation with bioinformatics specialists will be arranged to review data and findings CEG Protein Analysis F&S Core Proteomics Pilot Projects/Additional Workshops The Protein Analysis F&S Core is arranging to issue a request for new mini-pilot projects in proteomics. • First such project funded development of protein profiling (Michael Borchers) • Emphasis on projects requiring integration of sample preparation/data collection/data analysis • Expectation will be one project requiring existing capabilities and another project developing new skill sets within the core The Protein Analysis F&S Core will also be hosting several additional hands-on training and research-oriented workshops. Training/Workshop topics or ideas are welcome. CEG Protein Analysis F&S Core Contact Information Prof. Pat Limbach, Core Leader Pat.Limbach@uc.edu; 556-1871 Dr. Detlef Schumann, Director GRI Proteomics Lab schumad@ucmail.uc.edu; 558-8950 Prof. Michael Wagner, CCHMC, Proteomics Bioinformatics mwagner@cchmc.org; 636-2935 Dr. Yu Liang, Pharmacology, COM Proteomics Core liangyu@ucmail.uc.edu; 558-2347 Prof. Scott Belcher, Pharmacology, 2-D Gel Consultant belchesm@ucmail.uc.edu; 558-1721 Dr. Stephen Macha, UC Mass Spectrometry Facility, Proteomics Consultant machasf@email.uc.edu; 556-3950 CEG